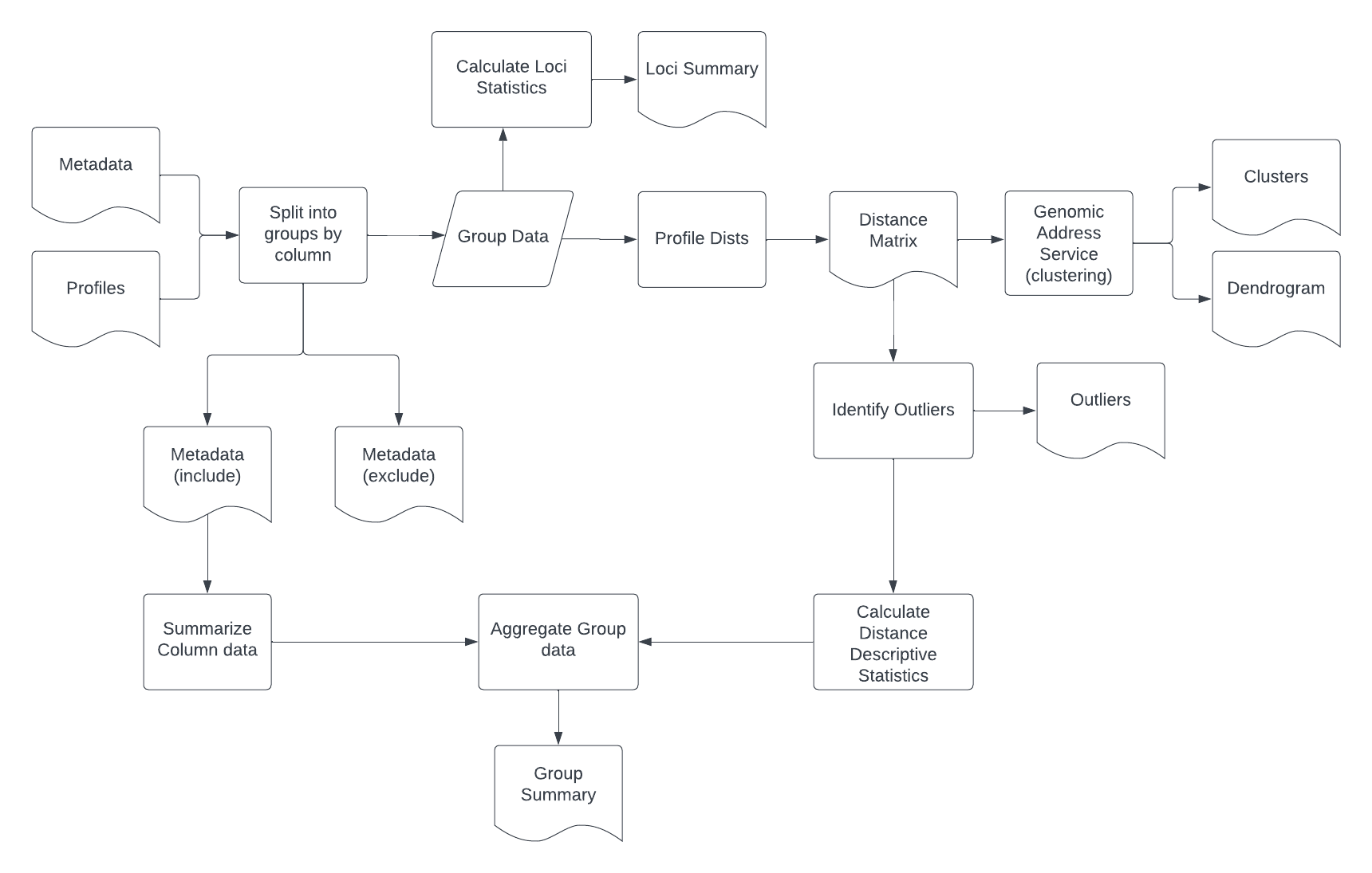
Opperationalized pathogen genomic surveillance and outbreak detection frequently makes use dendrograms in conjunction with organism specific genetic distance thresholds to "rule out" cases which are sufficiently genetically distinct that they are not part of the same "event". The genomic data is then combined with contextual sample data to assess the situation and inform further action.There are many different types of outbreaks within food/waterborne pathogens which may involve a general failure of a process where contamination is generalized and is multi-organism, which is identified through other means.
Arborator is designed to make the process of taking genomic profiles of alleles/snps/mutations and contextual metadata and perform:
- Splitting a large target collection of samples into invidual profile and metadata files
- Calculating within group genetic diversity statistics, generating dendrograms along with flat clusters based on thresholds, outlier detection, and loci summary reports
- Summarized report accross all analyzed groups
Install the latest released version from conda:
conda create -c bioconda -c conda-forge -n arborator arborator
Install using pip:
pip install arborator
Install the latest master branch version directly from Github:
pip install git+https://github.com/phac-nml/arborator.git
If you run arborator, you should see the following usage statement:
usage: arborator [-h] --profile PROFILE --metadata METADATA
[--config CONFIG] --outdir OUTDIR --partition_col
PARTITION_COL --id_col ID_COL
[--outlier_thresh OUTLIER_THRESH]
[--min_cluster_members MIN_CLUSTER_MEMBERS] [-n] [-s]
[--missing_thresh MISSING_THRESH] -t THRESHOLDS
[-d DELIMETER] [-e METHOD] [--force] [--cpus CPUS] [-V]
Run the test dataset using the data included in the repository under test_data
arborator --profile ./test_data/profile.tsv --metadata ./test_data/metadata.tsv --config ./test_data/config.json --outdir ./test_data/results --id_col id --partition_col outbreak --thresholds 10,9,8,7,6,5,4,3,2,1
Native
| id | locus_1 | locus_2 | locus_3 | locus_4 | locus_5 | locus_6 | locus_7 |
|---|---|---|---|---|---|---|---|
| S1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| S2 | 1 | 1 | 2 | 2 | ? | 4 | 1 |
| S3 | 1 | 2 | 2 | 2 | 1 | 5 | 1 |
| S4 | 1 | 2 | 3 | 2 | 1 | 6 | 1 |
| S5 | 1 | 2 | ? | 2 | 1 | 8 | 1 |
| S6 | 2 | 3 | 3 | - | ? | 9 | 0 |
- Direct support for missing data in the form of ?, 0, -, None or space
chewBBACA
| id | locus_1 | locus_2 | locus_3 | locus_4 | locus_5 | locus_6 | locus_7 |
|---|---|---|---|---|---|---|---|
| S1 | 1 | INF-2 | 1 | 1 | 1 | 1 | 1 |
| S2 | 1 | 1 | 2 | 2 | NIPH | 4 | 1 |
| S3 | 1 | 2 | 2 | 2 | 1 | 5 | 1 |
| S4 | 1 | LNF | 3 | 2 | 1 | 6 | 1 |
| S5 | 1 | 2 | ASM | 2 | 1 | 8 | 1 |
| S6 | 2 | INF-8 | 3 | PLOT3 | PLOT5 | 9 | NIPH |
- All non integer fields will be converted into missing data '0'
Hashes
| id | locus_1 | locus_2 | locus_3 | locus_4 | locus_5 | locus_6 | locus_7 |
|---|---|---|---|---|---|---|---|
| S1 | dc0a04880d1ad381ffd54ce9f6ad1e7a | - | b9f94bf167f34b9fcf45d79cab0e750a | 8a07b9cb0ab7560ad07b817ca34036bb | 80c8156d77d724ac0bb16ec60993bc84 | 7a1d0a48f16fa25910cddfea38dab229 | e1ee776b32c2f6131a7238ce50b75469 |
| S2 | dc0a04880d1ad381ffd54ce9f6ad1e7a | 0af06522a32865cd2db2cf5a854d195b | 9fc502308c616ae34146d7f7b0081bd8 | 4577dec2c840472800a3b104c88bb0ef | - | bba24c25c28c08058d6f32ecfbf509e9 | e1ee776b32c2f6131a7238ce50b75469 |
| S3 | dc0a04880d1ad381ffd54ce9f6ad1e7a | 04d45219ee5f6065caf426ba740215e5 | 9fc502308c616ae34146d7f7b0081bd8 | 4577dec2c840472800a3b104c88bb0ef | 80c8156d77d724ac0bb16ec60993bc84 | 874225c0dec5219dd64584ba32938dbd | e1ee776b32c2f6131a7238ce50b75469 |
| S4 | dc0a04880d1ad381ffd54ce9f6ad1e7a | - | e79562c280691c321612ecdf0dadad9e | 4577dec2c840472800a3b104c88bb0ef | 80c8156d77d724ac0bb16ec60993bc84 | c8087ad8b01d9f88e8eb2c3775ef2e64 | e1ee776b32c2f6131a7238ce50b75469 |
| S5 | dc0a04880d1ad381ffd54ce9f6ad1e7a | 04d45219ee5f6065caf426ba740215e5 | - | 4577dec2c840472800a3b104c88bb0ef | 80c8156d77d724ac0bb16ec60993bc84 | 4d547ea59e90173e8385005e706aae96 | e1ee776b32c2f6131a7238ce50b75469 |
| S6 | 8214e9d02d1b11e6239d6a55d4acd993 | - | e79562c280691c321612ecdf0dadad9e | - | - | e3088425be5e7de8d9a95da8e59a9ea8 | - |
- Direct support for missing data in the form of ?, 0, - or space
Native
| id | locus_1 | locus_2 | locus_3 | locus_4 | locus_5 | locus_6 | locus_7 |
|---|---|---|---|---|---|---|---|
| S1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| S2 | 1 | 1 | 2 | 2 | 0 | 4 | 1 |
| S3 | 1 | 2 | 2 | 2 | 1 | 5 | 1 |
| S4 | 1 | 2 | 3 | 2 | 1 | 6 | 1 |
| S5 | 1 | 2 | 0 | 2 | 1 | 8 | 1 |
| S6 | 2 | 3 | 3 | 0 | 0 | 9 | 0 |
- All columns are converted to contain only integers with missing data represented as a 0
Arborator allows for a great deal of flexibility with the input set of columns that can be supplied to the program. It has the constraints that the first column must be the sample identifier column, and it must be tab-delimited.
Example 1
| id | Country | Source | Date |
|---|---|---|---|
| S1 | Canada | Chicken | 2024-01-01 |
| S2 | Canada | Chicken | 2024-01-02 |
| S3 | United States | Chicken | 2024-01-03 |
| S4 | United Kingdom | Chicken | 2024-01-04 |
| S5 | Brazil | Chicken | 2023-12-01 |
| S6 | Canada | Chicken | 2023-11-02 |
Example 2
| sample_id | geo_loc | age | collection date | outbreak |
|---|---|---|---|---|
| S1 | Canada | 50 | 2024-01-01 | 1 |
| S2 | Canada | 25 | 2024-01-02 | 1 |
| S3 | United States | 10 | 2024-01-03 | 1 |
| S4 | United Kingdom | 1 | 2024-01-04 | 2 |
| S5 | Brazil | 56 | 2023-12-01 | 2 |
| S6 | Canada | 17 | 2023-11-02 | 2 |
There are a large number of parameters to configure within Arborator, so to enable consistency and ease for templating reports, we accept a configuration json object which allows the user to specify opperations for summarizing columns, and how they would be reported. Users can setup specific configurations for each of their target organisms of interest and use the config file as input to arborator for routine operations.
{
"outlier_thresh": "25",
"clustering_method": "average",
"clustering_threshold": "500,100,75,50,25,15,10,5,2,1,0",
"min_cluster_members": 2,
"partition_column_name": "outbreak",
"id_column_name": "sample_id",
"only_report_labeled_columns": "False",
"skip_qa": "False",
#Used to configure the order and opperations of columns in the grouped summary (optional)
"grouped_metadata_columns":{
"outbreak":{ "data_type": "None","label":"National Outbreak Code","default":"","display":"True"}, #Changing the label configures the output file to use this as the header name
"geo_loc":{ "data_type": "categorical","label":"Country of collection","default":"","display":"True"},
"age":{ "data_type": "desc_stats","label":"Patient Age (years)","default":"","display":"True"}, #Changing data_type to desc_stats causes the column to be reported in terms of min, median, mean, max values in the column
"collection date":{ "data_type": "min_max","label":"serovar","default":"","display":"True"}, #Changing data_type to min_mac causes the column to be reported in terms of min, max values in the column
},
#Used to configure the display of columns in the line list for individual samples (optional)
"linelist_columns":{
"outbreak":{ "data_type": "None","label":"National Outbreak Code","default":"","display":"True"},
"geo_loc":{ "data_type": "categorical","label":"organism","default":"","display":"True"},
"age":{ "data_type": "desc_stats","label":"Patient Age (years)","default":"","display":"True"}, #Changing data_type to desc_stats causes the column to be reported in terms of min, median, mean, max values in the column
"collection date":{ "data_type": "min_max","label":"serovar","default":"","display":"False"}, #Toggling false removes this column from output
}
}
Supported column summarization choices:
-
none - All values within the column are concatonated by a comma
-
categorical - Counts for all unique values are determined and then the count is provided in the format count_{column name}_{lable} : {count} (default)
-
min_max - The minimum and maximum values for the column are determined (dates and numerical data only) and reported as {column name}_min_value, {column name}_max_value,
-
desc_stats - Descriptive stats are determined for the column values (numerical data only) and reported as {column name}_min_value, {column name}_median_value, {column name}_mean_value, {column name}_max_value
Outputs:
{Output folder name}
├── {group label 1}
└── clusters.tsv
├── loci.summary.tsv
├── matrix.tsv
├── metadata.tsv
├── outliers.tsv
├── profile.tsv
└── tree.nwk
├── {group label n}
└── clusters.tsv
├── loci.summary.tsv
├── matrix.tsv
├── metadata.tsv
├── outliers.tsv
├── profile.tsv
└── tree.nwk
├── cluster_summary.tsv
├── metadata.excluded.tsv
├── metadata.included.tsv
├── threshold_map.json
└── run.json - Contains logging information for the run including parameters and quality information
Coming soon
Coming soon
Robertson, James, Wells, Matthew, Schonfeld, Justin, Reimer, Aleisha. Arborator: Streamlining public health pathogen outbreak and surveillance operations. 2024. https://github.com/phac-nml/arborator
Copyright Government of Canada 2023
Written by: National Microbiology Laboratory, Public Health Agency of Canada
Licensed under the Apache License, Version 2.0 (the "License"); you may not use this work except in compliance with the License. You may obtain a copy of the License at:
https://www.apache.org/licenses/LICENSE-2.0
Unless required by applicable law or agreed to in writing, software distributed under the License is distributed on an "AS IS" BASIS, WITHOUT WARRANTIES OR CONDITIONS OF ANY KIND, either express or implied. See the License for the specific language governing permissions and limitations under the License.
James Robertson: [email protected]