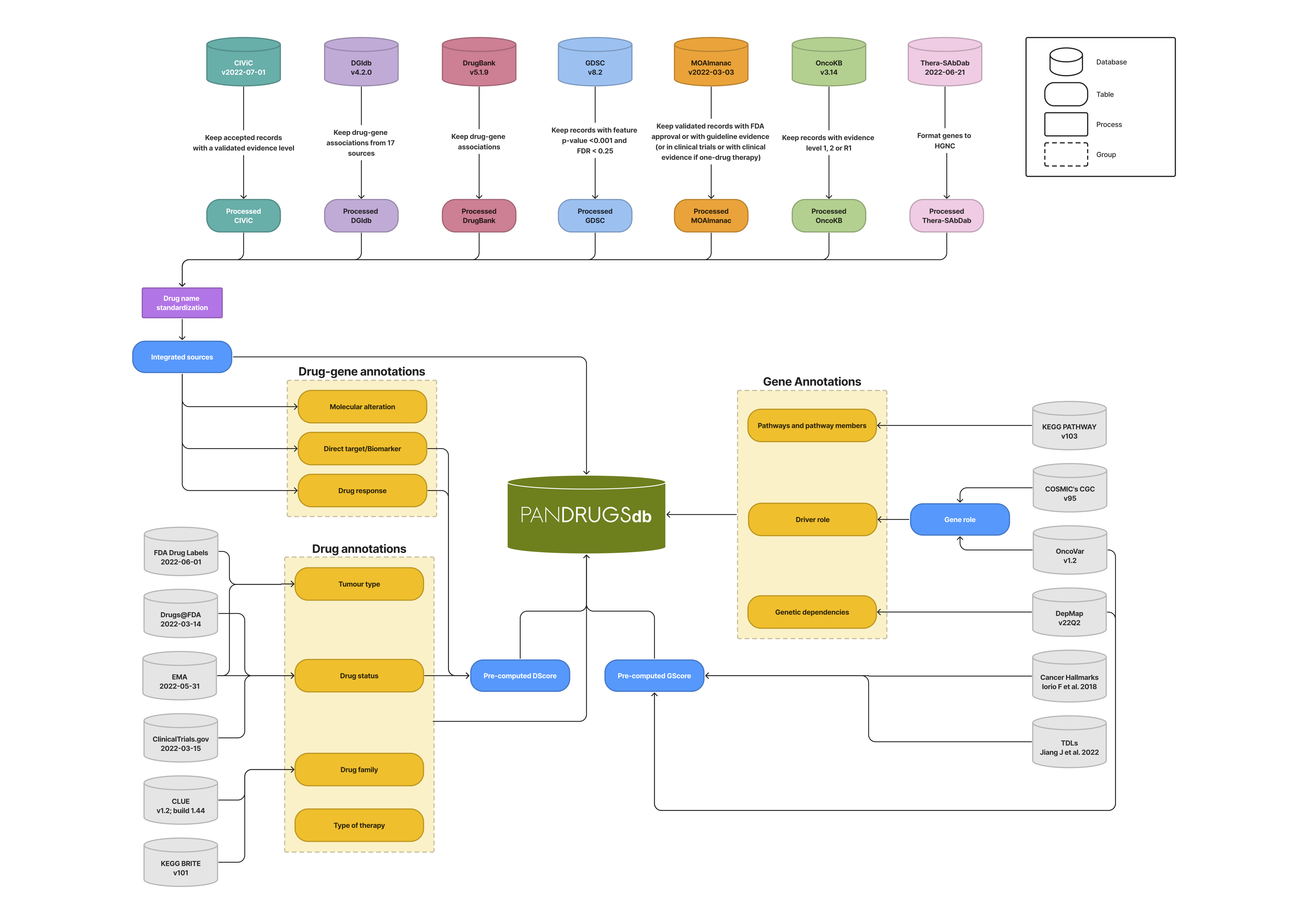
Pandrugs is a bioinformatics platform to prioritize anticancer drug treatments according to individual genomic data. Its database version 2.0 integrates data from 23 primary sources.
The code to generate its content and the annotations for VCF files is available in this github repository.
This workflow summarizes PanDrugsDB composition and the steps implemented for its construction.
In this table are described the sources included in the database and in the annotation process along with versions and license.
| Source | Version/Access Date*/DOI | License | Retrieved Information |
|---|---|---|---|
| CIViC | v2022-07-01 | CC0 1.0 | Drug-gene associations; Drug-gene annotations |
| DGIdb | v4.2.0 | MIT | Drug-gene associations |
| DrugBank | v5.1.9 | CC BY-NC 4.0 | Drug-gene associations |
| GDSC | v8.2 | CC BY-NC-ND 2.5 | Drug-gene associations; Drug-gene annotations |
| MOAlmanac | v2022-03-03 | GPLv2 | Drug-gene associations; Drug-gene annotations |
| OncoKB | v3.14 | Free for academic use | Drug-gene associations; Drug-gene annotations; Gene annotations |
| Thera-SAbDab | 2022-06-21 | CC BY 4.0 | Drug-gene associations |
| PubChem | 2022-11-29 | Free | Drug standardisation |
| HGNC | v2022-10-01 | CC0 1.0 | Gene standardisation |
| ClinicalTrials.gov | 2022-03-15 | Free | Drug annotations |
| CLUE | v1.2; build 1.44 | CC BY 4.0 | Drug annotations |
| Drugs@FDA | 2022-03-14 | Free | Drug annotations |
| EMA | 2022-05-31 | Free | Drug annotations |
| FDA Drug Labels | 2022-06-01 | CC0 1.0 | Drug annotations |
| KEGG BRITE | v101 | Free for academic use | Drug annotations |
| COSMIC's CGC | v95 | Free for non-commercial use | Gene annotations |
| DepMap | v22Q2 | CC BY 4.0 | Gene annotations; GScore calculation |
| KEGG PATHWAY | v103 | Free for academic use | Gene annotations; Variant annotations |
| OncoVar | v1.2 | Free for non-commercial use | Gene annotations; GScore calculation |
| Cancer Hallmarks | 10.1038/s41598-018-25076-6 | CC BY 4.0 | GScore calculation |
| TDLs | 10.1016/j.celrep.2022.110400 | CC BY-NC-ND 4.0 | GScore calculation |
| ClinVar | v2022-05 | Free | Variant annotations |
| COSMIC | v96 | Free for non-commercial use | Variant annotations |
| Domains | 10.1371/journal.pcbi.1004147 | CC BY 4.0 | Variant annotations |
| InterPro | v88.0 | CC0 1.0 | Variant annotations |
| Pfam | v35.0 | CC0 1.0 | Variant annotations |
| UniProt | v2022_01 | CC BY 4.0 | Variant annotations |
| VEP | v109 | Apache-2.0 | Variant annotations |
- Dates are displayed in ISO 8601 standard format: YYYY-MM-DD.
SL pairs are generated separately from the database and then merged back to the databse. The code responsible for generating and updating these pairs is stored here.