Welcome to PGxModule: Revolutionizing Genomic Medicine!
PGxModule is an advanced Nextflow DSL2 workflow, designed to seamlessly integrate into your genomics pipeline. It empowers you to generate sample-specific reports with clinical guidelines, leveraging state-of-the-art variant detection in Genomic Medicine Sweden sequencing panels. This workflow is inspired by JoelAAs. Besides, we have also implemented Pharmcat Pharmacogenomics Clinical Annotation Tool report with this pipeline, where we get recommendataion for all the detected haplotyopes directly from the CPIC.
- BAM-Centric Analysis: PGxModule is tailored to start directly from BAM files, streamlining the analysis process.
- Flexibility: Easily integrate into existing pipelines with analysis-ready BAM.
- Containerization: Supports Singularity for reproducible and efficient deployment.
- Customization: Tailor analyses with profiles such as HG38, HG19, Panel, WGS, and SOLID.
This pipeline branches into two analysis, one which only focuses on 19 SNPs from TPMT, DPYD, and NUDT15 genes, with plans to incorporate additional genes in future updates. The second part of the analysis is by using an external tool Pharmcat developed by PharmGKB where we try to find as many haplotypes as we can without subsetting the original bam, these haplotypes are then annotated and reported along with the clinical recommendations. The target selection is meticulously curated from reputable databases such as PharmGKB and PharmVar, guided by CPIC recommendations. As the pipeline evolves, it aims to broaden its scope, providing a more comprehensive analysis of pharmacogenomic variations to enhance clinical insights.
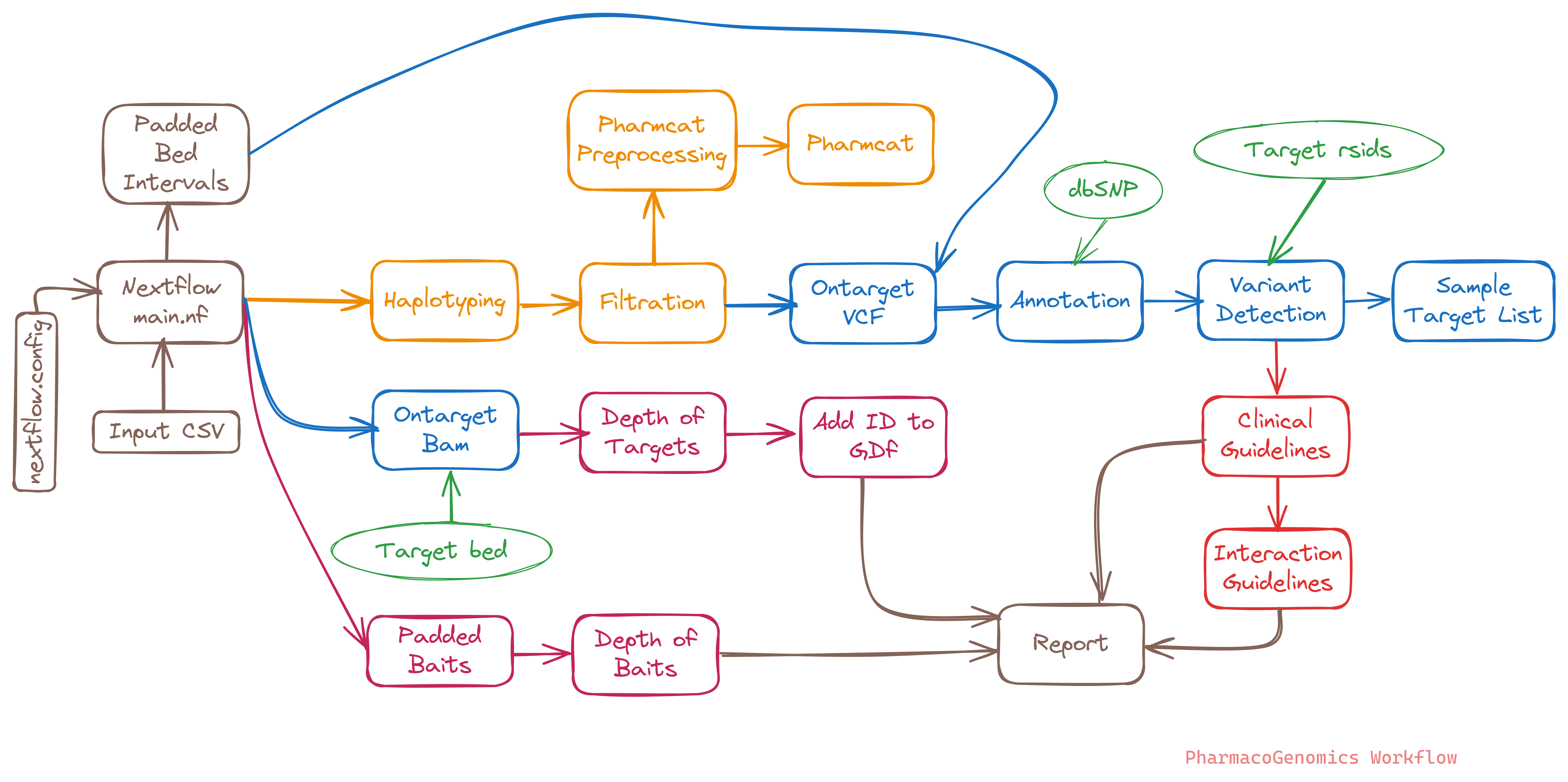
The PGxModule pipeline was executed with Nextflow version 23.04.2. The pipeline starts with the provided CSV file (configs/example.input.csv) containing sample information, initiating a comprehensive analysis tailored for pharmacogenomic studies.
- CSV Validation
The CSV Validation step ensures the correctness and integrity of the input CSV file. It checks for proper formatting, required fields, and data consistency, providing a foundation for accurate downstream processing in the PGxModule pipeline. - Haplotype Calling
Haplotype Calling is a crucial stage where the pipeline identifies and assembles haplotypes from the sequencing data. This process is fundamental in characterizing the genetic variations present in the samples, laying the groundwork for subsequent analyses and variant interpretation. - Haplotype Filtration
Haplotype Filtration focuses on refining the set of identified haplotypes, applying specific criteria to select variants of interest and discard noise. This process enhances the precision of the haplotype dataset, ensuring that downstream analyses are based on high-quality and clinically relevant variants. - PharmCat Preprocessing A script to preprocess VCF files for PharmCAT, ensuring compliance with VCF v4.2, stripping irrelevant PGx positions, normalizing variants, and optionally filtering sample data.
- PharmCat This scripts helps us to match the vcf positions with the pharmaco positions, runs the phenotypes and then finally the pharmcat report with all the recommendations.
- Ontarget VCF This step involves extracting the on-target VCF posiitons from the analyzed samples.
- Haplotype Annotation
Haplotypes which are called are annotated with dbSNP ids. - Detection of variants
Checking the variants of interest in the whole set of haplotypes and are used for futher analysis - Clinial Recommendations
Identified haplotypes are annotated with Haplotype Ids, clincial reccomendations, interaction guidelines based on CPIC. - Getting Ontarget Bam
This step involves extracting the on-target BAM files from the analyzed samples. These BAM files specifically capture the sequencing data aligned to the regions of interest, enabling reduction in time and focused analysis on the genomic regions relevant to the pharmacogenomic study. - Coverage Analysis
Coverage Analysis evaluates the sequencing depth across targeted regions, providing insights into the reliability of variant calls. By assessing coverage, this step identifies regions with insufficient data and informs the overall confidence in the accuracy of the genomic information obtained from the samples. - Report
The Report step consolidates the findings from the preceding analyses into a comprehensive report. This report includes detailed information on detected variants, clinical guidelines, interaction assessments, and other relevant pharmacogenomic insights. It serves as a valuable resource for clinicians and researchers, aiding in informed decision-making based on the genomic characteristics of the analyzed samples.
| clarity_sample_id | id | type | assay | group | bam | bai | purity |
|---|---|---|---|---|---|---|---|
| XXX000001 | Sample1 | T | gmssolidpgx | Sample1 | Sample1.T.bwa.umi.sort.bam | Sample1.T.bwa.umi.sort.bam.bai | 0.30 |
| XXX000002 | Sample2 | T | gmssolidpgx | Sample2 | Sample2.T.bwa.umi.sort.bam | Sample2.T.bwa.umi.sort.bam.bai | 0.30 |
- Nextflow version >= 23.04.0
- Singularity version >= 3.8.0
For instructions on installing Nextflow, refer to the Nextflow documentation for installation guidance based on your operating system.
Singularity is used in conjunction with Nextflow for containerization. You can install Singularity by following the installation instructions provided on the Singularity website.
After installing Singularity, make sure it is added to your system's PATH to enable seamless integration with Nextflow.
To set up the workflow, configure the paths in configs/nextflow.config. If using Singularity with Nextflow, run the script envs/get_containers.sh.
To utilize Sentieon, follow these steps:
- Ensure that the Sentieon executable is in the system's path.
- If needed, store license information as a secret in Nextflow's local store.
- Set the environmental variable
NXF_ENABLE_SECRETSto an appropriate value.
nextflow secrets set SENTIEON_LICENSE_BASE64 <LICENSE>If using Nextflow secrets, set the environment variable NXF_ENABLE_SECRETS to true. This ensures that the pipeline can retrieve the secret from Nextflow's secrets store during execution. Note that versions of Nextflow from 22.09.2-edge onwards have NXF_ENABLE_SECRETS set to true by default. If not using secrets, set NXF_ENABLE_SECRETS to false, but ensure that the environment variable SENTIEON_LICENSE reflects the value of your license server on your machine.
- HG38
- HG19 (soon to be depreicated)
- Panel
- WGS
- SOLID
nextflow run main.nf --csv /path/to/csv/input.csv -profile "panel,hg38,solid" --haplotype_caller SENTIEON |& tee logs/run.log