WO2021094805A1 - Resistance to acidovorax valerianellae in corn salad - Google Patents
Resistance to acidovorax valerianellae in corn salad Download PDFInfo
- Publication number
- WO2021094805A1 WO2021094805A1 PCT/IB2019/001290 IB2019001290W WO2021094805A1 WO 2021094805 A1 WO2021094805 A1 WO 2021094805A1 IB 2019001290 W IB2019001290 W IB 2019001290W WO 2021094805 A1 WO2021094805 A1 WO 2021094805A1
- Authority
- WO
- WIPO (PCT)
- Prior art keywords
- plant
- qtl
- valerianellae
- resistance
- allele
- Prior art date
Links
Classifications
-
- A—HUMAN NECESSITIES
- A01—AGRICULTURE; FORESTRY; ANIMAL HUSBANDRY; HUNTING; TRAPPING; FISHING
- A01H—NEW PLANTS OR NON-TRANSGENIC PROCESSES FOR OBTAINING THEM; PLANT REPRODUCTION BY TISSUE CULTURE TECHNIQUES
- A01H5/00—Angiosperms, i.e. flowering plants, characterised by their plant parts; Angiosperms characterised otherwise than by their botanic taxonomy
- A01H5/12—Leaves
-
- A—HUMAN NECESSITIES
- A01—AGRICULTURE; FORESTRY; ANIMAL HUSBANDRY; HUNTING; TRAPPING; FISHING
- A01H—NEW PLANTS OR NON-TRANSGENIC PROCESSES FOR OBTAINING THEM; PLANT REPRODUCTION BY TISSUE CULTURE TECHNIQUES
- A01H6/00—Angiosperms, i.e. flowering plants, characterised by their botanic taxonomy
Definitions
- the present invention relates to resistance, especially intermediate resistance, in corn salad plants, namely plants of Valerianella locusta or Valerianella locusta L, to bacterial spot disease caused by the bacteria Acidovorax valerianellae.
- the resistance is provided by DNA sequences, or QTL, introgressed into the genome of Valerianella locusta plants.
- the introgressed sequences can be present homozygously or heterozygously in the genome of a Valerianella locusta L. plant.
- the present invention also relates to corn salads, comprising the DNA sequences or QTL that lead to resistance to A. valerianellae.
- the invention further relates to markers linked to said DNA sequences and to the use of such makers to identify or select the DNA sequences and to identify or select plants carrying such resistance.
- the invention also relates to the seeds and progeny of such plants and to propagation material for obtaining such plants, and to different uses of these plants.
- Corn salad breeding relies mainly on phenotypic selection (Muminovic J. et al. 2004). According to the standard criteria of International Union for the Protection of New Varieties of Plants (UPOV), Corn salad varieties are described and differentiated using morphological traits such as seed size and shape, leaf length, shape, profile, glossiness, color, thickness, and prominence of veins. Genetic studies were limited to amplified fragment length polymorphism (AFLP) fingerprinting to estimate the genetic diversity of corn salad germplasm (Vos P. et al. 1995; Muminovic J. et al. 2004). Conventional breeding aims at producing varieties with round and dark green leaves and is successful in developing such plant cultivars and germplasm. Yet, developing disease resistances is still costly and difficult, especially because of poor genomic resources dedicated to this species.
- AFLP amplified fragment length polymorphism
- Bacterial black spot caused by Acidovorax valerianellae (Gardan L. et al., 2003) is a major disease of corn salad in France and is responsible for about 10% losses (in tonnage) on average per year. It was first observed in 1991 , first localized in western France, in high technology field production for the preparation of ready to use salads. It is now spread more globally in all production areas (including Belgium, Germany and Austria). Its occurrence increases with the number of successive crops without break between crop (Grondeau & Samson, 2009). Black and greasy spots on leaves and cotyledons reduce corn-salad quality significantly, and the harvest of infected crops is unmarketable when the number of spotted plants exceeds 5-10%. The spots could also be framed by a yellow halo and progressively, the whole leaf could become yellow.
- A. valerianellae in corn salad would be beneficial in the control of the black spot disease. Such resistance would not only increase the stability of crop protection, but would also result in a reduced, or eliminated, requirement for environmentally harmful, and not always effective, fungicide applications. However, about 30 years after the first identification of the disease, no genetic control of the A. valerianellae is available for V. locusta L. production.
- the present inventors have identified a resistance against the A. valerianellae pathogen in a wild Valerianella locusta f. carinata (also known as Valerianella carinata) plant and have been able to introgress this resistance into V. locusta L. plants, thus obtaining resistant V. locusta L. plants.
- the resistance of the present invention is imparted by the newly discovered sequences conferring the resistance, said resistance being linked to an additive quantitative trait loci (QTL), transferable to different V. locusta L genetic backgrounds.
- the present invention provides cultivated corn salads that display resistance against the A. valerianellae pathogen as well as methods that produce or identify corn salads that exhibit resistance against A. valerianellae.
- the present invention also discloses molecular genetic markers, especially Single Nucleotide Polymorphisms (SNPs), linked to the QTL that lead to resistance against the A. valerianellae pathogen. Plants obtained through the methods and uses of such molecular markers are also provided.
- Said resistance is moreover easily transferable to different genetic backgrounds and the invention also extends to different methods allowing the transfer or introgression of the QTL conferring the phenotype.
- the invention also provides several methods and uses of the information linked to the SNPs associated to the QTL conferring A. valerianellae resistance, inter alia methods for identifying A. valerianellae resistant plants and methods for identifying further molecular markers linked to this resistance to bacterial spot disease caused by A. valerianellae , as well as methods for improving the yield of corn salad production in an environment infected by A. valerianellae and methods for protecting a corn salad culture from A. valerianellae infection or transmission and for controlling the back spot disease caused by A. valerianellae. Definitions:
- the term “Resistance” is as defined by the ISF (International Seed Federation) Vegetable and Ornamental Crops Section for describing the reaction of plants to pests or pathogens, and abiotic stresses for the Vegetable Seed Industry.
- resistance it is meant the ability of a plant variety to restrict the growth and/or development of a specified pest or pathogen and/or the damage it causes when compared to susceptible plant varieties under similar environmental conditions and pest or pathogen pressure. Resistant varieties may exhibit some disease symptoms or damage under heavy pest or pathogen pressure. Two levels of resistance are defined.
- High resistance plant varieties that highly restrict the growth and/or development of the specified pest or pathogen and/or the damage it causes under normal pest pressure when compared to susceptible varieties. These plant varieties may, however, exhibit some symptoms or damage under heavy pest/pathogen pressure.
- Intermediate resistance plant varieties that restrict the growth and/or development of the specified pest or pathogen and/or the damage it causes but may exhibit a greater range of symptoms or damage compared to high resistant varieties. Intermediate resistant plant varieties will still show less severe symptoms or damage than susceptible plant varieties when grown under similar environmental conditions and/or pest pressure.
- A. valerianellae infection The disease symptoms of A. valerianellae infection are black and greasy spots on leaves and cotyledons, rendering the salad unmarketable.
- the spots could also be framed by a yellow halo and progressively, the whole leaf could become yellow.
- Susceptibility is the inability of a plant variety to restrict the growth and/or development of a specified pest or pathogen.
- a susceptible plant thus displays the detrimental symptoms linked to the bacterial infection, namely the foliar damages.
- an offspring plant refers to any plant resulting as progeny from a vegetative or sexual reproduction from one or more parent plants or descendants thereof.
- an offspring plant may be obtained by cloning or selfing of a parent plant or by crossing two parental plants and include selfings as well as the F1 or F2 or still further generations.
- An F1 is a first-generation offspring produced from parents at least one of which is used for the first time as donor of a trait, while offspring of second generation (F2) or subsequent generations (F3, F4, etc.) are specimens produced from selfings of FTs, F2's etc.
- An F1 may thus be (and usually is) a hybrid resulting from a cross between two true breeding parents (true-breeding is homozygous for a trait), while an F2 may be (and usually is) an offspring resulting from self-pollination of said F1 hybrids.
- cross refers to the process by which the pollen of one flower on one plant is applied (artificially or naturally) to the ovule (stigma) of a flower on another plant.
- the term “genetic determinant” and/or “QTL” refers to any segment of DNA associated with a biological function.
- QTLs and/or genetic determinants include, but are not limited to, genes, coding sequences and/or the regulatory sequences required for their expression.
- QTLs and/or genetic determinants can also include nonexpressed DNA segments that, for example, form recognition sequences for other proteins.
- the term “genotype” refers to the genetic makeup of an individual cell, cell culture, tissue, organism (e.g., a plant), or group of organisms.
- heterozygote refers to a diploid or polyploid individual cell or plant having different alleles (forms of a given gene, genetic determinant or sequences) present at least at one locus.
- heterozygous refers to the presence of different alleles (forms of a given gene, genetic determinant or sequences) at a particular locus.
- homozygote refers to an individual cell or plant having the same alleles at one or more loci on all homologous chromosomes.
- homozygous refers to the presence of identical alleles at one or more loci in homologous chromosomal segments.
- hybrid refers to any individual cell, tissue or plant resulting from a cross between parents that differ in one or more genes.
- locus refers to any site that has been defined genetically, this can be a single position (nucleotide) or a chromosomal or genomic region.
- a locus may be a gene, a genetic determinant, or part of a gene, or a DNA sequence, and may be occupied by different sequences.
- a locus may also be defined by a SNP (Single Nucleotide Polymorphism), by several SNPs, or by two flanking SNPs.
- the invention encompasses plants of different ploidy levels, essentially diploid plants, but also triploid plants, tetraploid plants, etc.
- DNA strand and allele designation and orientation for the SNP markers is done according to the TOP/BOT method developed by lllumina (https://www.illumina.com/documents/products/technotes/technote_topbot.pdf).
- the present inventors have identified a QTL which, when present in Valerianella locusta L. plants, provides resistance, essentially intermediate resistance to these plants when infected or likely to be infected by the A. valerianellae bacteria.
- the seeds and plants according to the invention have been obtained from an initial cross between a wild plant of Valerianella carinata or Valerianella locusta f. carinata plant, the introgression partner displaying the phenotype of interest, and a plant of Valerianella locusta L, the recurrent susceptible parent, in order to transfer the resistance into Valerianella locusta L. genetic background.
- the present invention is thus directed to a corn salad plant or seed, which is resistant against A. valerianellae , comprising in its genome a quantitative trait locus (QTL), conferring said resistance against. A. valerianellae.
- QTL quantitative trait locus
- Said QTL conferring the resistance was initially introgressed from a wild V. locusta f. carinata, also known as V. carinata plant and are thus referred to as the resistance QTL or introgressed sequences of the invention in the following description.
- the invention is also directed to a cell of such a plant or seed, comprising these introgressed sequences conferring the resistance.
- the resistance phenotype can be tested and scored as described in example 1 of the experimental section, by natural infection or by artificial inoculation for example by vaporization.
- the resistance against A. valerianellae according to the invention is best characterized by intermediate resistance.
- the introgressed sequences conferring the resistance are preferably identified with the SNP (Single Nucleotide Polymorphism) molecular marker CS-0022531 (SEQ ID NO:14).
- SNP Single Nucleotide Polymorphism
- This SNP can thus be used as a marker of the presence or absence of the resistance QTL or introgressed sequences.
- the presence of the introgressed sequences, or QTL, conferring the resistance phenotype is identified by allele A of SNP marker CS-0022531.
- a SNP refers to a single nucleotide in the genome, which is variable depending on the allele which is present, whereas the flanking nucleotides are identical.
- the SNPs are disclosed in table 2 also by reference to theirflanking sequences, identified by SEQ ID number.
- sequence associated with a specific SNP in the present application for example SEQ ID NO:14 for the SNP CS-0022531
- only one nucleotide within the sequence actually corresponds to the polymorphism namely the 151 st nucleotide of SEQ ID NO:14 corresponds to the polymorphic position of SNP CS- 0022531 , which can be A or G as indicated in table 2.
- the flanking sequences are given for positioning the SNP in the genome but are not part of the polymorphism as such.
- the inventors have moreover shown that the presence of the QTL conferring the resistance phenotype of the invention can also be tested by detection of a specific haplotype, namely a specific combination of alleles for two SNPs, which combination is informative and representative of the presence of the sequences conferring the resistance.
- This haplotype is preferably the haplotype corresponding to allele A of SNP CS-0022531 (SEQ ID NO: 14) in combination with allele A of SNP CS-0015357 (SEQ ID NO:9). Consequently, the presence of the QTL conferring the resistance phenotype can be tested or confirmed by the detection of the specific combination of allele A of CS- 0022531 and allele A of CS-0015357.
- the QTL according to the invention and conferring the intermediate resistance against A. valerianellae is preferably chosen from the ones present in the genome of seeds of ValeriacidoR. A sample of this Valerianella locusta seed has been deposited by HM. Clause S.A. Rue Louis Saillant, Z.l.
- a QTL of the invention is thus preferably identical to introgressed sequences from V. carinata present in the genome of the seeds of ValeriacidoR, NCIMB 43500.
- the present inventors have also localized the QTL/introgressed sequences responsible for the phenotype of interest.
- the QTL has not been localized with respect to the chromosomes of V. locusta, but on the basis of the linkage groups, as identified by the inventors, using classical genotyping approaches and tools described in example 3.
- the QTL responsible for the phenotype of interest is to be found within a genomic region or interval which comprises or is delimited by SNP CS-0019663 (SEQ ID NO:7) and SNP CS-0021333 (SEQ ID NO: 10), preferably in the region comprising or delimited by SNP CS-0019663 (SEQ ID NO:7) and SNP CS-0015357 (SEQ ID NO:9), and even more preferably in the region comprising or delimited by SNP CS-0016322 (SEQ ID NO:8) and SNP CS-0015357 (SEQ ID NO:9). All these SNP are to be found on Linkage Group 3 (LG3) as identified by the inventors; they are also found on the same contig, and thus are on the same chromosome.
- LG3 Linkage Group 3
- a genomic region identified by flanking sequences e.g. SNPs, is defined clearly and non- ambiguously.
- the QTL, or introgressed sequences from V. carinata are located within a genomic interval delimited by CS-16322 (SEQ ID NO:8) and CS-0015357 (SEQ ID NO:9), or overlaps with said interval.
- a genomic region delimited by two SNPs X and Y refers to the section of the genome, more specifically of a chromosome, lying between the positions of these two SNPs and preferably comprising said SNPs, therefore the nucleotide sequence of this chromosomal region begins with the nucleotide corresponding to SNP X and ends with the nucleotide corresponding to SNP Y, i.e. the SNPs are comprised within the region they delimit, according to the invention.
- genomic region under test has the same sequence, in the sense of the invention, as the corresponding genomic region found in the V. carinata donor at the same locus, if said genomic region under test is also capable of conferring resistance to A. valerianellae.
- GISH genetic in situ hybridization
- a corn salad plant, cell or seed of the invention is thus preferably also homozygous for the QTL, or introgressed sequences of the invention conferring A. valerianellae resistance.
- the invention is however not limited to such homozygous plants, cells or seeds. Indeed, the inventors have also demonstrated that an intermediate resistance is also present in plants having heterozygously the QTL of interest (see experimental section).
- the present invention thus also encompasses corn salad plant, cell or seed having heterozygously in their genome the QTL, or introgressed sequences, of the invention as defined above.
- the inventors have identified and mapped the QTL imparting the intermediate resistance of the invention, mainly by identifying the presence of sequences representative of the introgressed QTL at different loci, namely at least at 5 different loci defined by the 5 following SNPs: CS-0019663 (SEQ ID NO:7), CS-0016322 (SEQ ID NO:8), CS-0015357 (SEQ ID NO:9), CS-0021333 (SEQ ID NO:10) and CS-0022531 (SEQ ID NO:14).
- the alleles of these molecular markers representative of the QTL or introgressed sequences conferring the resistance of the invention are allele A of CS-0019663, allele A of CS-0016322, allele A of CS-0015357, allele A of CS-0021333 and allele A of CS-0022531.
- the presence of the QTL in the genome of a corn salad plant, cell or seed according to the invention can thus be detected or revealed by detecting sequences representative of the QTL at said loci, more preferably by detecting one or more of the following alleles of the corresponding molecular markers: allele A of CS-0019663, allele A of CS-0016322, allele A of CS-0015357, allele A of CS-0021333 and allele A of CS-0022531.
- the QTL of the invention conferring resistance against A. valerianellae can be detected in a plant, cell, seed, or any other plant part by one or more of the following molecular markers: CS- 0022531 , CS-0015357 and/or CS-0016322. More preferably, said QTL is detected by any one of allele A of CS-0016322, allele A of CS-0015357 and/or allele A of CS-0022531.
- the QTL confers intermediate resistance when present at the homozygous and heterozygous states
- its presence can thus be detected in a plant, cell, seed or any plant part, by detecting one or more of the following genotypes of markers CS-0022531 , CS-0015357 and/or CS- 0016322:
- a plant, seed or cell of the invention is thus characterized by at least one of the above genotypes, preferably by the genotype AA or AG of CS-0022531 , or by one of the genotypes (AA, AA), (AA, AG), (AG, AA) and (AG, AG) for (CS-0022531 , CS-0015357).
- a corn salad plant, cell or seed according to the invention is a Valerianella locusta L. plant, a Valerianella locusta L. locusta plant or a Valerianella locusta plant.
- a corn salad plant, cell or seed according to the invention is not from the wild species Valerianella locusta L. carinata, or Valerianella carinata.
- a corn salad plant of the invention is a cultivated plant or line, more preferably an edible, commercial, cultivated corn salad plant.
- a commercial plant or line may also exhibit resistance to other viruses or pests.
- the corn salad plant, cell or seed, or other plant part, of the invention also comprises another type of resistance or intermediate resistance to A. valerianellae , which is not conferred by the introgressed sequences of the invention, but by any other gene or QTL conferring said resistance or intermediate resistance, in order to provide high resistance to the plant, cell or seed, or other plant part, of the invention.
- the invention is directed to corn salad plants, exhibiting resistance against bacterial spot disease caused by A. valerianellae bacteria, as well as to seeds giving rise to those plants, and cells of these plants or seeds, or other plant parts, comprising the resistance QTL in their genome, introgressed from V. carinata.
- the invention encompasses plants, seeds and cells of any ploidy levels, it encompasses inter alia diploid, triploid, tetraploid and/or allopolyploid plants, cells or seeds.
- a plant or seed according to the invention may be a progeny or offspring of a plant grown from the deposited seeds ValeriacidoR, deposited at the NCIMB under the accession number NCIMB 43500. Plants grown from the deposited seeds are indeed homozygous for the QTL of the invention conferring the intermediate resistance of interest, they thus bear in their genome the QTL of interest homozygously. They can be used to transfer these introgressed sequences into another background by crossing and selfing and/or backcrossing.
- the invention is also directed to the deposited seeds of ValeriacidoR (NCIMB 43500) and to plants grown from one of these seeds. These seeds contain homozygously the QTL conferring the phenotype of interest.
- the invention is also directed to plants or seeds as defined above, i.e. containing the resistance QTL of interest in homozygous or heterozygous state, wherein these plants or seeds are obtainable by transferring the QTL from a Valerianella locusta L. plant, representative seeds thereof were deposited under NCIMB accession NCIMB 43500, into another Valerianella locusta L. genetic background, for example by crossing said deposited plant with a second corn salad parent and selection of the plant or plants bearing the QTL responsible for the phenotype of interest. During such crossing, the QTL can be transferred.
- seeds or plants of the invention may also be obtained by different processes not necessarily implying crossing and introgression, and are not exclusively obtained by means of an essentially biological process.
- the invention relates to a cultivated corn salad plant or seed, preferably a non-naturally occurring corn salad plant or seed, which may comprise one or more mutations in its genome, which provides the plant with resistance against A. valerianellae , which mutation is as present, for example, in the genome of plants of which a representative sample was deposited with the NCIMB under deposit number NCIMB 43500.
- the invention in another embodiment, relates to a method for obtaining a corn salad plant or seed carrying one or more mutations in its genome, which provides the plant with resistance against A. valerianellae.
- a method for obtaining a corn salad plant or seed carrying one or more mutations in its genome which provides the plant with resistance against A. valerianellae.
- Such a method is illustrated in example 6 and may comprise: a) treating M0 seeds of a corn salad to be modified with a mutagenic agent to obtain M1 seeds; b) growing plants from the thus obtained M1 seeds to obtain M1 plants; c) producing M2 seeds by self-fertilization of M1 plants; and d) optionally repeating step b) and c) n times to obtain M1+n seeds.
- the M1+n seeds are grown into plants and submitted to A. valerianellae infection.
- the surviving plants, or those with the milder symptoms of A. valerianellae infection, are multiplied one or more further generations while continuing to be selected for their resistance against A. valerianellae.
- the M1 seeds of step a) can be obtained via chemical mutagenesis such as EMS mutagenesis.
- chemical mutagenic agents include but are not limited to, diethyl sufate (des), ethyleneimine (ei), propane sultone, N-methyl-N-nitrosourethane (mnu), N-nitroso-N-methylurea (NMU), N-ethyl-N-nitrosourea(enu), and sodium azide.
- the mutations are induced by means of irradiation, which is for example selected from x-rays, fast neutrons, UV radiation.
- the mutations are induced by means of genetic engineering.
- Such mutations also include the integration of sequences conferring the resistance against A. valerianellae , as well as the substitution of residing sequences by alternative sequences conferring the resistance against A. valerianellae.
- the mutations are the integration of the QTL as described above, in replacement of the homologous sequences of a V. locusta L. plant.
- the mutations are the substitutions of the sequences comprised within SNP CS-0019663 and SNP CS-0021333 of the Valerianella locusta L.
- the genetic engineering means which can be used include the use of all such techniques called New Breeding Techniques which are various new technologies developed and/or used to create new characteristics in plants through genetic variation, the aim being targeted mutagenesis, targeted introduction of new genes or gene silencing (RdDM).
- New Breeding Techniques which are various new technologies developed and/or used to create new characteristics in plants through genetic variation, the aim being targeted mutagenesis, targeted introduction of new genes or gene silencing (RdDM).
- Example of such new breeding techniques are targeted sequence changes facilitated through the use of Zinc finger nuclease (ZFN) technology (ZFN-1 , ZFN-2 and ZFN-3, see U.S. Pat. No.
- Oligonucleotide directed mutagenesis ODM
- Cisgenesis Cisgenesis and intragenesis
- Grafting on GM rootstock
- Reverse breeding Agro-infiltration
- agro-infiltration agro-infiltration "sensu stricto", agro-inoculation, floral dip
- Transcription Activator-Like Effector Nucleases TALENs, see U.S. Pat. Nos. 8,586,363 and 9,181 ,535)
- the CRISPR/Cas system see U.S. Pat. Nos.
- Such applications can be utilized to generate mutations (e.g., targeted mutations or precise native gene editing) as well as precise insertion of genes (e.g., cisgenes, intragenes, or transgenes).
- the applications leading to mutations are often identified as site-directed nuclease (SDN) technology, such as SDN1 , SDN2 and SDN3.
- SDN1 site-directed nuclease
- SDN1 site-directed nuclease
- a SDN is used to generate a targeted DSB and a DNA repair template (a short DNA sequence identical to the targeted DSB DNA sequence except for one or a few nucleotide changes) is used to repair the DSB: this results in a targeted and predetermined point mutation in the desired gene of interest.
- the SDN3 is used along with a DNA repair template that contains new DNA sequence (e.g. gene).
- the outcome of the technology would be the integration of that DNA sequence into the plant genome.
- the most likely application illustrating the use of SDN3 would be the insertion of cisgenic, intragenic, or transgenic expression cassettes at a selected genome location.
- JRC Joint Research Center
- the resistance, or intermediate resistance, against A. valerianellae corresponds, according to the invention, to a reduction of at least 20% of the symptoms of A. valerianellae infection on leaves and cotyledons, at around 10 to 20 days after infection, with respect to the symptoms on leaves and cotyledons of a plant devoid of said QTL, preferably a reduction of at least 40%, 50%, 60%, 70% or more of said symptoms.
- the score rate of a plant of the invention is at least 7 in the scale described in example 1 , i.e. 25% or less of symptoms, preferably 12% or less of symptoms, corresponding to a score of 8 or more, and preferably a score of “9” corresponding to a symptomless plant, in the conditions of infection described in example 1.
- the invention in another aspect also concerns any plant likely to be obtained from seed or plants of the invention as described above, and also plant parts of such a plant, and most preferably explant, scion, cutting, seed, fruit, root, rootstock, pollen, ovule, embryo, protoplast, leaf, anther, stem, petiole, and any other plant part such as cell, wherein said plant, explant, scion, cutting, seed, fruit, root, rootstock, pollen, ovule, embryo, protoplast, leaf, anther, stem, petiole, and/or plant part is obtainable from a seed or plant according to the first aspect of the invention, i.e. bearing the QTL of interest, also called introgressed sequences of the invention, in their genome.
- These plant parts inter alia explant, scion, cutting, seed, fruit, root, rootstock, pollen, ovule, embryo, protoplast, leaf, anther, stem or petiole, comprise in their genome the QTL conferring the phenotype of interest, i.e. resistance against A. valerianellae ; they thus comprise a cell comprising this resistance QTL or introgressed sequences from V. carinata.
- this QTL or introgressed sequences is/are preferably as found in the genome of the deposited seed ValeriacidoR NCIMB 43500.
- the invention is directed to seed as described above, which develops into a plant according to the first aspect of the invention, thus resistant against A. valerianellae infection thanks to the presence of the resistance QTL, or introgressed sequences, as defined above.
- the invention is also directed to cells of V. locusta plants, such that these cells comprise, in their genome, the introgressed QTL of the present invention conferring resistance against A. valerianellae to a V. locusta plant.
- the QTL is as already defined in the context of the present invention, it is characterized by the same features and preferred embodiments already disclosed with respect to the plants and seeds according to the preceding aspects of the invention.
- the presence of this QTL, or introgressed sequences can be revealed by the techniques disclosed above and well known to the skilled reader.
- the QTL can inter alia be identified in the genome of such a cell of the invention with the molecular marker CS-0022531 , especially allele A of this marker. It is advantageously characterized by the presence of the haplotype corresponding to allele A of CS-0022531 and allele A of CS-0015357. Said QTL can be present homozygously or heterozygously.
- Cells according to the invention can be any type of V. locusta L. or V. locusta cell, inter alia an isolated cell and/or a cell capable of regenerating a whole corn salad, bearing the QTL or introgressed sequences of interest.
- the cell is not a cell of the wild species V. carinata.
- the present invention is also directed to a tissue culture of non-regenerable or regenerable cells of the plant as defined above according to the present invention; preferably, the regenerable or non- regenerable cells are derived from embryos, protoplasts, meristematic cells, callus, pollen, leaves, anthers, stems, petioles, roots, root tips, fruits, seeds, flowers, cotyledons, and/or hypocotyls of the invention, and the cells contain the QTL, or introgressed sequences, of interest, homozygously or heterozygously in their genome conferring resistance against A. valerianellae.
- the tissue culture will preferably be capable of regenerating plants having the physiological and morphological characteristics of the foregoing corn salad, and of regenerating plants having substantially the same genotype as the foregoing corn salad plant.
- the present invention also provides corn salad plants regenerated from the tissue cultures of the invention.
- the invention also provides a protoplast of the plant defined above, or from the tissue culture defined above, said protoplast containing the QTL, or introgressed sequences, conferring the resistance phenotype of the invention.
- the invention is also directed to tissue of a plant of the invention; the tissue can be an undifferentiated tissue, or a differentiated tissue.
- tissue comprises one or more cells comprising the QTL of the invention.
- the invention is also directed to propagation material, capable of producing a resistant corn salad according to the invention, comprising the introgressed sequences or QTL as defined above.
- the invention also provides food product comprising the corn salad of the invention, or parts thereof, especially leaves or young shoots, which comprise cells having the introgressed QTL of the invention in their genome.
- the present invention is also directed to the use of a corn salad plant of the invention, preferably comprising homozygously the QTL of the invention, as a breeding partner in a breeding program for obtaining V. locusta plants, preferably edible corn salad plants having resistance against A. valerianellae , namely resistance against bacterial spot disease caused by A. valerianellae.
- a breeding partner harbors, preferably homozygously in its genome the QTL, or introgressed sequences, conferring the resistance of interest.
- a plant according to the invention, or any part thereof comprising the QTL of interest can thus be used as a breeding partner for introgressing the QTL, or introgressed sequences, conferring the desired resistance into an edible corn salad or V. locusta L. germplasm and/or for breeding V. locusta plants resistant against A. valerianellae infection.
- a plant or seed bearing the QTL of interest heterozygously can also be used as a breeding partner, the segregation of the QTL, or introgressed sequences, is likely to render the breeding program more complex.
- the improved phenotype of the invention is resistance against A. valerianellae , namely resistance against bacterial spot disease caused by A. valerianellae. It is preferably to be transferred into a plant which is initially susceptible to A. valerianellae infection. As already mentioned above, it can also be transferred into a plant comprising another form of A. valerianellae resistance, conferred by another QTL, gene or introgression.
- the introgressed QTL will advantageously be introduced into varieties or OP varieties that contain other desirable genetic traits such as resistance to others diseases or pest, good agronomical features, drought tolerance, and the like.
- the invention is also directed to the same use with plants or seed of ValeriacidoR, deposited at the NCIMB under the accession number NCIMB 43500. Said plants are also suitable as introgression partners in a breeding program aiming at conferring the desired phenotype to V. locusta plant or germplasm.
- the selection of the progeny displaying the desired phenotype, or bearing the QTL linked to resistance against A. valerianellae can advantageously be carried out on the basis of the alleles of the SNP markers, especially the SNP markers of the invention, and preferably SNP markers CS-0022531 , CS-0015357 and/or CS-0016322, more preferably the alleles of the haplotype constituted by CS-0022531 and CS-0015357, and even more preferably by CS- 0022531.
- a progeny is preferably selected on the presence of at least one of the following alleles: allele A of CS-0019663, allele A of CS-0016322, allele A of CS-0015357, allele A of CS-0021333 and allele A of CS-0022531.
- the progeny is advantageously selected on the basis of the presence of allele A of CS-0022531 , or the combination of allele A of CS-0022531 and allele A of CS-0015357.
- the progeny is preferably selected on the presence of one of these alleles homozygously.
- a plant according to the invention or grown from a seed as deposited under accession number NCIMB 43500, is thus particularly valuable in a marker assisted selection for obtaining commercial corn salad lines and varieties, having the improved phenotype of the invention, namely resistance against A. valerianellae , more specifically intermediate resistance.
- the invention is also directed to the use of said plants in a program aiming at identifying, sequencing and / or cloning the genetic introgressed sequences conferring the desired phenotype.
- the invention also concerns methods or processes for the production or breeding of V. locusta plants, especially edible cultivated corn salads, having the desired phenotype, especially commercial plants and inbred parental lines.
- the present invention is indeed also directed to transferring the QTL, or introgressed sequences of the invention conferring the resistance against A. valerianellae , to other corn salads, especially other corn salad varieties, or other species or inbred parental lines, and is useful for producing new types and varieties of corn salad.
- the invention also comprises methods for breeding V. locusta plants having resistance against A. valerianellae , comprising the steps of crossing a plant grown from the deposited seeds ValeriacidoR NCIMB 43500 or progeny thereof bearing the QTL of the invention conferring A. valerianellae resistance, with an initial V. locusta plant preferably devoid of said QTL.
- the QTL is as defined above, namely introgressed from V. carinata and is preferably present in the genome of the seeds of ValeriacidoR, NCIMB accession number 43500.
- This QTL is identifiable by CS-0022531 , preferably by allele A of said SNP, and also preferably by the haplotype corresponding to allele A of CS-0022531 and allele A of CS-0015357.
- the invention also concerns a method for conferring resistance against A. valerianellae to a V. locusta plant, comprising genetically modifying said plant to introduce the resistance QTL of the invention.
- Said QTL is as defined above and is preferably present in the genome of the seeds of ValeriacidoR, NCIMB accession number 43500.
- the genetic modification can be carried out by any methods or means well known to the skilled person.
- the invention is also directed to a method or process for the production of a plant having resistance against A. valerianellae; such a method or process may comprise the following steps: a) Crossing a plant grown from a deposited seed NCIMB 43500, or progeny thereof comprising the QTL conferring resistance against A. valerianellae , and an initial V. locusta plant, preferably devoid of said QTL, b) Selecting one plant in the progeny thus obtained, bearing the QTL of the present invention; c) Optionally self-pollinating one or several times the plant obtained at step b) and selecting in the progeny thus obtained a plant having A. valerianellae resistance, especially intermediate resistance.
- the method or process may comprise instead of step a) the following steps: a1) Crossing a plant corresponding to the deposited seeds (NCIMB 43500), or progeny thereof, bearing the QTL conferring A. valerianellae resistance, and an initial V. locusta plant, preferably devoid of said QTL, a2) Increasing the F1 hybrid by means of selfing to create F2 population.
- SNP markers are preferably used in steps b) and / or c), for selecting plants bearing sequences conferring the resistance phenotype of interest.
- the SNP markers are preferably one or more of the 5 preferred SNP markers of the invention, namely CS-0019663, CS-0016322, CS-0015357, CS-0021333 and CS-0022531 , including all combinations thereof, especially any combination comprising SNP CS-0022531 , preferably comprising CS- 0015357 and CS-0022531 , inter alia the combination of the three markers CS-0016322, CS-0015357 and CS-0022531.
- the plant is selected as having A. valerianellae resistance, intermediate resistance or improved resistance with respect to the initial plant, when the allele of the SNP(s) is (are) the allele corresponding to the allele of the resistant parent for this SNP and not the allele of the initial susceptible V. locusta plant.
- Those alleles for the SNPs of the invention have already been detailed in this application and are reported in table 2.
- a plant can be selected as having the improved phenotype of the invention, when allele A of CS-0019663, allele A of CS-0016322, allele A of CS-0015357, allele A of CS-0021333 and/or allele A of CS-0022531 is detected.
- the V. locusta plant of step a) is an elite line, used in order to obtain a plant with commercially desired traits or desired horticultural traits.
- a method or process as defined above may advantageously comprise backcrossing steps, preferably after step c), in order to obtain plants having all the characterizing features of V. locusta plants, especially of edible corn salad. Consequently, a method or process for the production of a plant having these features may also comprise the following additional steps: d) Backcrossing the resistant plant selected in step b) or c) with a V. locusta plant; e) Selecting a plant bearing the QTL, or introgressed sequences, of the present invention.
- the plant used in step a), namely the plant corresponding to the deposited seeds can be a plant grown from the deposited seeds; it may alternatively be any plant according to the 1 st aspect of the invention, bearing the QTL or introgressed sequences conferring the phenotype.
- SNP markers can be used for selecting plants having an improved resistance to A. valerianellae , with respect to the initial plant.
- the SNP markers are those of the invention, as described in the previous sections.
- the method or process of the invention is carried out such that, for at least one of the selection steps, namely b), c) and/or e), the selection is based on the detection of at least one of the resistance alleles corresponding to allele A of CS-0019663, allele A of CS- 0016322, allele A of CS-0015357, allele A of CS-0021333 and allele A of CS-0022531 , preferably detection of allele A of CS-0022531 or detection of allele A of CS-0015357 and allele A of CS- 0022531.
- the preferred alleles and combinations have already been disclosed and are applicable to this embodiment.
- the presence of the preferred alleles or combinations is informative regarding the presence of the QTL of the invention, conferring the phenotype of interest.
- plants having the improved phenotype, and bearing homozygously the QTL, or introgressed sequences, of the invention are to be selected, the selection is to be made advantageously on the basis of one or more the SNPs of the invention, on the presence of the alleles representative of the QTL, namely the alleles found in the resistant parent, in combination with the absence of the alleles representative of the recurrent susceptible V. locusta parent.
- the selection can also be made on the basis of any other marker linked to the introgressed sequences and representative of the presence of these introgressed sequences by opposition to the resident sequences of the susceptible parent. Methods for defining alternative markers are also in the scope of the present invention.
- the plant selected at step e) is preferably a cultivated or commercial corn salad plant.
- steps d) and e) are repeated at least twice and preferably three times, not necessarily with the same V. locusta plant.
- Said V. locusta plant(s) is (are) preferably a breeding line (s).
- the self-pollination and backcrossing steps may be carried out in any order and can be intercalated, for example a backcross can be carried out before and after one or several self-pollinations, and self- pollinations can be envisaged before and after one or several backcrosses.
- the method used for allele detection can be based on any technique allowing the distinction between two different alleles of a SNP or other marker, at a defined genomic locus.
- the present invention also concerns a plant obtained or obtainable by such a method.
- a plant is indeed a V. locusta plant having the resistance phenotype according to the first aspect of the invention.
- the invention is also directed to a method for obtaining cultivated or commercial corn salad plants or inbred parental lines thereof, having the desired improved phenotype, corresponding to an improved resistance to A. valerianellae with respect to an initial commercial V. locusta plant, comprising the steps of: a) Backcrossing a plant obtained by germinating a deposited seed ValeriacidoR NCIMB accession number 43500, or progeny thereof, bearing the QTL conferring A. valerianellae resistance, with a commercial corn salad plant, b) Selecting a plant bearing the QTL.
- the selection is made on the basis of one or more of the 5 SNPs of the invention, as detailed for the other methods of the invention, preferably a selection comprising at least selection of allele A of CS-0022531 , and preferably selection of allele A of CS-0015357 and allele A of CS- 0022531.
- Alternative marker of the QTL can also be used.
- the present invention is also directed to a V. locusta plant and seed obtainable by any of the methods and processes disclosed above.
- the seed of such V. locusta are preferably coated or pelleted with individual or combined active species such as plant nutrients, enhancing microorganisms, or products for disinfecting the environment of the seeds and plants.
- active species such as plant nutrients, enhancing microorganisms, or products for disinfecting the environment of the seeds and plants.
- Such species and chemicals may be a product that promotes the growth of plants, for example hormones, or that increases their resistance to environmental stresses, for example defense stimulators, or that stabilizes the pH of the substrate and its immediate surroundings, or alternatively a nutrient.
- viruses and pathogenic microorganisms for example a fungicidal, bactericidal, hematicidal, insecticidal or herbicidal product, which acts by contact, ingestion or gaseous diffusion; it is, for example, any suitable essential oil, for example extract of thyme. All these products reinforce the resistance reactions of the plant, and/or disinfect or regulate the environment of said plant.
- They may also be a live biological material, for example a nonpathogenic microorganism, for example at least one fungus, or a bacterium, or a virus, if necessary with a medium ensuring its viability; and this microorganism, for example of the pseudomonas, bacillus, trichoderma, clonostachys, fusarium, rhizoctonia, etc. type stimulates the growth of the plant, or protects it against pathogens.
- a live biological material for example a nonpathogenic microorganism, for example at least one fungus, or a bacterium, or a virus, if necessary with a medium ensuring its viability
- this microorganism for example of the pseudomonas, bacillus, trichoderma, clonostachys, fusarium, rhizoctonia, etc. type stimulates the growth of the plant, or protects it against pathogens.
- the identification of the plants bearing the QTL, or introgressed sequences, responsible for the improved resistance to A. valerianellae could be done by the detection of at least one of the alleles of the SNPs associated with the introgressed resistance QTL, potentially in combination with the absence of the other allelic form of the SNPs of the present invention, in order to confirm the homozygous state of the QTL if needed.
- the identification of a plant bearing homozygously the QTL or introgressed sequences of the present invention will be based on the identification of at least one of allele A of CS-0019663, allele A of CS-0016322, allele A of CS-0015357, allele A of CS-0021333 and allele A of CS-0022531 , as well as the absence of the other allele for these SNPs, which are as disclosed in table 2.
- the invention is also directed to the use of the information provided herewith by the present inventors, namely the existence of a QTL, or introgressed sequences, present in the deposited seeds of ValeriacidoR (NCIMB accession number 43500), and conferring the resistance phenotype to V. locusta plants, and the disclosure of molecular markers associated to this QTL or introgressed sequences.
- This knowledge can be used inter alia for precisely mapping the QTL, for defining its sequence, for identifying corn salad plants comprising the QTL conferring the resistance phenotype and for identifying further or alternative markers associated to this QTL.
- Such further markers are characterized by their location, namely close to the 5 markers disclosed in the present invention, preferably close to at least one of the 2 markers of the haplotype, and by their association with the phenotype of interest, as revealed by the invention, namely A. valerianellae resistance, more specifically intermediate resistance against this pathogen.
- the invention is thus also directed to a method for identifying, detecting and/or selecting V. locusta plants having the QTL of the present invention as found in the genome of the seeds of ValeriacidoR (NCIMB accession number 43500), said QTL conferring an improved A.
- valerianellae resistance with respect to a corresponding plant devoid of said QTL, the method comprising the detection of at least one of the following markers: allele A of CS-0019663, allele A of CS-0016322, allele A of CS- 0015357, allele A of CS-0021333 and allele A of CS-0022531 , preferably allele A of CS-0022531 potentially in combination with allele A of CS-0015357, in a genetic material sample of the plant to be identified and/or selected.
- the invention is also directed to a method for detecting or selecting V. locusta plants having the QTL conferring A. valerianellae resistance and having at least one of the alleles mentioned above, wherein the detection or selection is made on condition of A. valerianellae infection comprising inoculation of A. valerianellae on the plants to be tested, for example inoculation by vaporization.
- the presence of the phenotype of interest is informative of the presence of the QTL or introgressed sequences of the invention.
- the invention is also directed to a method for detecting and/or selecting a V. locusta plant, especially cultivated edible corn salad, having the QTL of the invention, comprising the detection of at least one of the alleles mentioned above.
- the method is particularly adapted in a breeding program with ValeriacidoR (NCIMB accession number 43500), as initial parent, or progeny thereof, comprising the QTL of the invention conferring A. valerianellae resistance.
- the invention is further directed to a method for detecting and/or selecting V. locusta plants, especially cultivated edible corn salad, having the QTL of the present invention conferring the resistance phenotype, on the basis of the detection of any molecular marker revealing the presence of said QTL or informative about the presence of said QTL.
- the QTL may be characterized by the presence of at least one of the SNPs of the invention, especially CS-0022531 , or by the haplotype corresponding to CS-0022531 and CS-0015357, but it may also be identified through the use of different, alternative markers. Also included in the present invention are thus methods and uses of any such molecular markers for identifying the QTL of the invention in a corn salad genome, wherein said QTL confers an improved A.
- valerianellae resistance with respect to a corresponding plant devoid of said QTL, the QTL being characterized by the presence of at least one of allele A of CS-0019663, allele A of CS-0016322 , allele A of CS-0015357 , allele A of CS-0021333 and allele A of CS-0022531 , preferably at least by allele A of CS-0022531.
- any such alternative molecular marker for identifying the QTL or introgressed sequences of the invention in a corn salad genome confers A. valerianellae resistance and is characterized by the presence of at least one of allele A of CS- 0019663, allele A of CS-0016322, allele A of CS-0015357, allele A of CS-0021333 and allele A of CS-0022531 , preferably at least by allele A of CS-0022531.
- the invention also concerns a method for detecting and/or selecting corn salad plants having the resistance QTL as defined previously, conferring A. valerianellae resistance, said method comprising: a) Assaying corn salad plants for the presence of at least one genetic marker genetically linked or associated to the QTL involved in A. valerianellae resistance, b) Selecting a plant comprising the genetic marker and the linked or associated QTL involved in A. valerianellae resistance, wherein the QTL and the genetic marker are to be found in the genomic region delimited by CS- 0019663 and CS-0021333 in the genome of V. locusta.
- association or genetic association, and more specifically genetic linkage, it is to be understood that a genetic polymorphism of the marker (e.g. a specific allele of the SNP marker) and the phenotype of interest occur simultaneously, i.e. are inherited together, more often than would be expected by chance occurrence, i.e. there is a non-random association of the allele and of the genetic sequences responsible for the phenotype, as a result of their genomic proximity.
- the marker e.g. a specific allele of the SNP marker
- the phenotype of interest occur simultaneously, i.e. are inherited together, more often than would be expected by chance occurrence, i.e. there is a non-random association of the allele and of the genetic sequences responsible for the phenotype, as a result of their genomic proximity.
- a molecular marker of the invention is inherited with the phenotype of interest in preferably more than 90% of the meioses, preferably in more than 95%, 96%, 98% or 99% of the meioses.
- the invention thus concerns the use of one or more molecular markers, for fine-mapping or identifying a QTL in the corn salad genome, said QTL participating to the improved phenotype of the invention, wherein said one or more markers is/are localized in one of the following genomic regions:
- said one or more markers is in the genomic region delimited by CS-0016322 and CS-0021333, preferably by CS-0016322 and CS-0015357, more preferably by CS-0022531 and CS-0015357.
- Said one or more molecular marker(s) is/are moreover preferably associated, with a p-value of 0.05 or less, with at least one of the following SNP alleles: allele A of CS-0019663, allele A of CS-0016322, allele A of CS-0015357, allele A of CS-0021333 and allele A of CS-0022531.
- the molecular marker is preferably a SNP marker. It is more preferably at less than 1 megabase from the locus of at least one of the 5 SNPs of the invention, preferably of one of the 3 markers CS- 0016322, CS-0015357 and CS-0022531 , and preferably of CS-0022531.
- the QTL is to be found in the deposited seeds NCIMB 43500.
- the p-value is preferably less than 0.01.
- the invention is also directed to the use of at least one of the 5 SNPs of the invention, associated with the QTL conferring A. valerianellae resistance to corn salad, for identifying one or more alternative molecular markers associated with said QTL, wherein said one or more alternative molecular markers are:
- said one or more markers are in the genomic region delimited by CS-0016322 and CS-0021333, preferably by CS-0016322 and CS-0015357, more preferably by CS-0022531 and CS-0015357.
- the genetic association or linkage can advantageously be detected by following the alternative maker and the presence of the QTL in the progeny arising from a plant comprising the QTL of interest.
- the alternative molecular markers are preferably associated with said QTL with a p-value of 0.05 or less, preferably less than 0.01. The QTL is to be found in the deposited seeds NCIMB 43500.
- the QTL mentioned above, conferring A. valerianellae resistance according to the invention, is the introgressed QTL present in ValeriacidoR (NCIMB accession number 43500).
- association or linkage is as defined above; preferably the association or linkage is with a p- value of preferably less than 0.05, and most preferably less than 0.01 or even less.
- a molecular marker and the resistance phenotype are inherited together in preferably more than 90% of the meioses, preferably more than 95%.
- the molecular markers according to this aspect of the invention are most preferably SNP. They are more preferably at less than 1 megabase from the locus of at least one of the 5 SNPs of the invention, preferably at less than 0.5 megabases.
- the invention also comprises a method for identifying a molecular marker associated with a QTL participating to A. valerianellae resistance in corn salad, as described in the present application, comprising the steps of:
- CS-0019663 and CS- 0021333 identifying a molecular marker in the genomic region delimited by CS-0019663 and CS- 0021333, or at less than 2 megabase units from the locus of one of CS-0016322, CS-0022531 and CS-0015357, preferably less than 1 or less than 0.5 megabase units;
- the invention is also directed to the use of a molecular marker for identifying or selecting a corn salad plant comprising, in its genome, a QTL conferring to V. locusta plants A. valerianellae resistance, wherein said marker is in the genomic region delimited by CS-0019663 and CS-0021333, or at less than 2 megabase unit from the locus of at least one of the 5 SNP markers of the invention; and wherein said molecular marker is associated with at least one of the following SNP alleles: allele A of CS-0019663, allele A of CS-0016322, allele A of CS-0015357, allele A of CS-0021333 and allele A of CS-0022531 , with a p-value of 0.05 or less, preferably 0.01 or less.
- the molecular marker to be used according to this embodiment is obtainable inter alia by the method for identifying further or alternative molecular markers, as disclosed above.
- the molecular marker is preferably a SNP marker. It is more preferably at less than 1 megabase from the locus of at least one of the 5 SNPs of the invention.
- the invention is also directed to a method for genotyping a plant, preferably a V. locusta plant or corn salad germplasm, for the presence of at least one genetic marker associated with A. valerianellae resistance, wherein the method comprises the determination or detection in the genome of the tested plant of a nucleic acid comprising at least one of the markers of the invention, or comprising at least one of the alternative molecular markers as disclosed above.
- the method comprises the step of identifying in a sample of the plant to be tested specific sequences associated with A. valerianellae resistance, in nucleic acids comprising at least one of the alleles of the SNPs of the invention, as already disclosed.
- the detection of a specific allele of a SNP can be carried out by any method well known to the skilled reader.
- the resistant plants of the invention In view of the ability of the resistant plants of the invention to restrict the damages caused by A. valerianellae infection, namely to restrict bacterial spot disease caused by A. valerianellae bacteria, they are advantageously grown in an environment infected or likely to be infected by A. valerianellae ; in these conditions, the resistant plants of the invention produce more marketable corn salad than susceptible plants.
- the invention is thus also directed to a method for improving the yield of corn salad in an environment infected by A. valerianellae comprising growing corn salads comprising in their genome a QTL, or introgressed sequences, as defined according to the previous aspects of the invention, and conferring to said plants A. valerianellae resistance.
- the method comprises a first step of choosing or selecting a corn salad plant having said QTL, or introgressed sequences, of interest.
- the method can also be defined as a method of increasing the productivity of a corn salad field, tunnel, greenhouse or glasshouse, or as a method of reducing the intensity or number of fungicide application in the production of corn salad.
- the invention is also directed to a method for reducing the loss on corn salad production in conditions of A. valerianellae infection, especially loss due to unmarketable corn salads, comprising growing a corn salad plant as defined above.
- said methods for improving the yield or reducing the loss on corn salad production may comprise a first step of identifying corn salad plants resistant against A. valerianellae resistance and comprising in their genome a QTL that confers to said plants A. valerianellae resistance, and then growing said resistant plants in an environment infected or likely to be infected by the bacteria.
- the resistant plants of the invention are also able to restrict at least partially, the multiplication of A. valerianellae , thus limiting the infection of further plants and the propagation of the bacteria.
- the invention is also directed to a method of protecting a field, tunnel, greenhouse or glasshouse, or any other type of plantation, from A. valerianellae infection, or of at least limiting the level of infection by A. valerianellae of said field, tunnel, greenhouse or glasshouse or of limiting the spread of A. valerianellae in a field, tunnel, greenhouse or glasshouse, especially in glasshouse.
- Such a method preferably comprises the step of growing a resistant plant of the invention, i.e. a plant comprising in its genome a QTL conferring to said plant A. valerianellae resistance.
- the invention also concerns the use of a plant resistant to A. valerianellae for controlling A. valerianellae infection, or controlling the black spot disease caused by A. valerianellae bacteria, in a field, tunnel, greenhouse or glasshouse, or other plantation; such a plant is a plant of the invention, comprising in its genome a QTL as defined above. According to this use, the plants of the invention are therefore used for protecting a field, tunnel, greenhouse or glasshouse from A. valerianellae infection.
- All the preferred features of the QTL are as defined in connection with the other aspects of the invention, namely it is present in the seeds of ValeriacidoR (NCIMB accession number 43500), and it is identifiable by the SNP marker CS-0022531 , preferably by allele A of CS-0022531 , or identifiable by the haplotype corresponding to allele A of CS-0022531 and allele A of CS-0015357.
- FIG. 1 Distribution of the Acidovorax score in an F2 population (152 different plants) of SRLIxSusceptible Parent; the X-axis represents the Acidovorax score according to the rating disclosed in example 1. The Y-axis represent the number of progenies.
- FIG.2 Genomic positions of the QTLs detected on the Linkage Group 3 (LG 3) for F2 population of -SRL1 x Susceptible”
- FIG.3 Genomic position of the QTL detected on the Linkage Group 3 (LG 3) of ‘“SRL1 x Susceptible” map by multiple QTL mapping (MQM) for the Acidovorax resistance score noted on leaves.
- LG 3 Linkage Group 3
- MQM multiple QTL mapping
- X-axis represents the genomic positions of the SNPs along the linkage group 3.
- the Y-axis represent the LOD Score.
- FIG.4 Distribution of Acidovorax scores obtained within the panel of the 74 inbred lines tested for validation; the scores illustrated are the mean of two different measures.
- FIG.5 Boxplot of adjusted values of resistance to Acidovorax in the 74 inbred lines, as a function of the genotypes at QTL3 haplotype consisting of SNP markers CS-0015357 and CS-0022531 (RR: Resistant alleles (homozygously) for both SNPs, RS: Heterozygous state for both SNPs, i.e. presence of the Resistant allele and of the susceptible allele for CS-0015357 and CS-0022531 , SS: Susceptible alleles (homozygously) for both SNPs).
- RR Resistant alleles (homozygously) for both SNPs
- RS Heterozygous state for both SNPs, i.e. presence of the Resistant allele and of the susceptible allele for CS-0015357 and CS-0022531
- SS Susceptible alleles (homozygously) for both SNPs).
- FIG.6 Results of analyses of variance of QTL3 on Acidovorax level of resistance.
- the meaning of RR, RS and SS is as defined for FIG. 5.
- a French strain of Acidovorax valerianellae has been collected in France area.
- the inoculum is prepared by multiplying the bacteria on solid King B medium for 48 hours at 26°C and further diluted at 10 8 bacteria per ml in NaCI 0.9% solution (D.O. 0.3 at 600 nm).
- Plantlets to be tested are grown in greenhouses for 3 weeks (3-4 leaves old plantlets stage). They are cultivated at 20-24°C in batches of 25 seeds of the genotype to be tested by repetition (total 6 reps). Plantlets are inoculated by vaporization and grown in growth chambers at 21 °C.
- Example 2 Identification of a resistant plant.
- the present inventors identified a wild Valerianella locusta F. carinata (SRL1) showing some intermediate resistance to Acidovorax valerianellae in pathology test set up according to the protocol described in example 1. This accession was further tested five times and always scored among the best accessions, displaying a strong Acidovorax score according to the test described in Example 1.
- the SRL1 resistant plant was crossed with a variety of Valerianella locusta L. susceptible to Acidovorax valerianellae, and the F1 was further selfed, thereby creating an F2 population of 152 different plants to be evaluated according to the pathology test of example 1.
- susceptible and resistant seeds of the parents were planted as controls.
- the distribution of the Acidovorax score is represented in FIG. 1 for the studied progenies; the average score for the studied progenies and its parental lines is reported in table 1.
- Table 1 Average Acidovorax score for the studied progenies and its parental lines.
- the normal distribution (see Fig. 1) tends to indicate that the resistance behaves as a complex trait relying on a polygenic genetic architecture, rather than a single gene model.
- HM. Clause variety GALA
- HM. Clause was fully sequenced and assembled in order to obtain a reference sequence ( de novo whole genome sequencing approach - WGS).
- GGS genotyping by sequencing approach
- 48 F2 plants from the population established in example 2 were analyzed in order to identify the specific variations existing between the parental material of the said population, especially the ones linked to the Acidovorax valerianellae resistance.
- genotyping by sequencing approach was performed according to the protocol described in Elshire RJ et al., 2011 for the libraries preparation, followed by a next generation lllumina HiSeq sequencing for the genotyping by sequencing itself.
- the whole genome sequencing assembly was done using PacBio long read technology (Rhoads A, Au KF, 2015).
- the Map construction was carried out using JoinMap version 4.0 (JoinMap 4, Software for the calculation of genetic linkage maps in experimental populations. Kyasma BV, Wageningen, Netherlands). Chi square values were calculated for each marker to detect segregation deviations (P ⁇ 0.05) from the expected Mendelian ratio and highly distorted markers were discarded.
- LG Linkage groups
- a first step was the Kruskal-Wallis analysis in which, the effect of the marker significance level on a specific trait, here the Acidovorax valerianellae resistance was estimated by simple linear regression between the marker and the trait (Kenis K, Keulemans J, 2007).
- QTL mapping was performed using interval mapping (IM) which estimates the likelihood score of a putative QTL placed in any position within an interval flanked by two adjacent markers (Jansen RC and Stam P, 1994).
- IM interval mapping
- the LOD score threshold (P-value of 0.05) at which a QTL was declared significant, was determined using a permutation test for each LG as well as for the genome wide. Over 1000 permutations the frequency distribution of the maximum LOD score under the null hypothesis (no QTL) was determined (Churchill GA, Doerge RW 1994).
- markers When several markers displayed a LOD score equals or superior to the LOD score determined by the permutation test, they were declared as cofactors for a multiple QTL mapping (MQM) analysis with a step size of 1 cM (Jansen RC and Stamp P, 1994).
- MQM multiple QTL mapping
- linkage group 3 for Acidovorax valerianellae resistance.
- the linkage group 3 is created and its positions attributed by the mapping software, independently from the Valerianella locusta L. chromosomes or of their physical positions on said chromosomes.
- QTL3 This QTL on LG3 is also referred to as “QTL3” in the following.
- KASPar assay used for validation was preformed based on KASP method from KBioscience (LGC Group, Teddington, Middlesex, UK).
- KASP SNP assays were designed using LGC's primer picker software. For every SNP, two allele-specific forward primers and one common reverse primer per SNP assay were designed. KASP genotyping assays are based on competitive allele-specific PCR and enable bi- allelic scoring of SNPs at specific loci. To summarize, the SNP-specific KASP assay mix and the universal KASP Master mix were added to DNA samples, a thermal cycling reaction was then performed, followed by an end-point fluorescent read.
- Biallelic discrimination was achieved through the competitive binding of two allele-specific forward primers, each with a unique tail sequence that corresponded with two universal FRET (fluorescence resonant energy transfer) cassettes, one of which was labelled with FAMTM dye and the other of which was labelled with VICTM dye (LGC, www.lgcgroup. com).
- FRET fluorescence resonant energy transfer
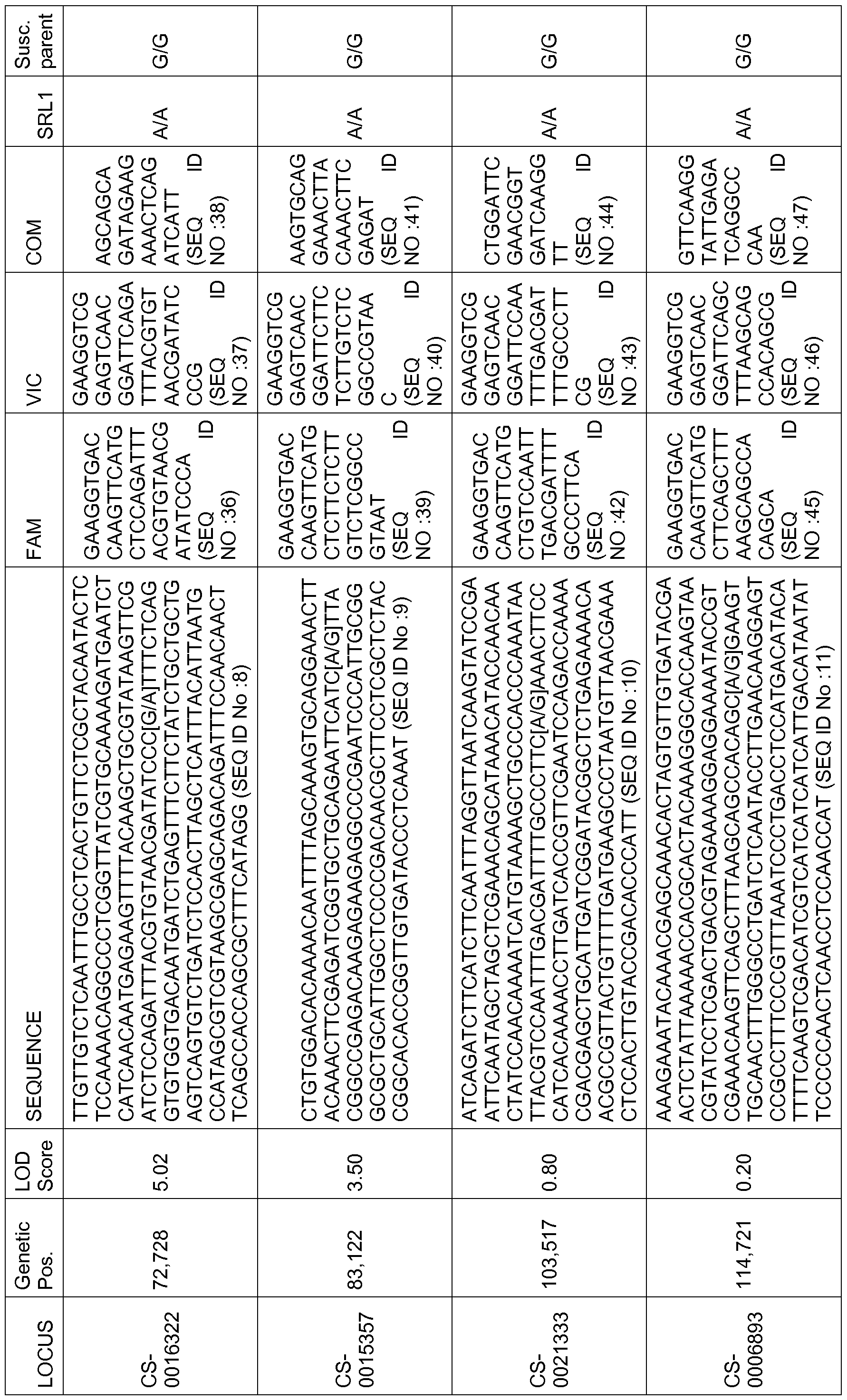
- SNPs sequences of the SNPs, including flanking sequences, of the two allele-specific forward primers, of the reverse primer, as well as the allele representative of the susceptible and resistant parent, are given in table 2.
- able 2 Sequences of the SNPs. Column 1 reports the name of the SNP, column 2 indicates the position (Pos.) on LG3, column 3 indicates the LOD score, column 4 the sequence of the bi allelic SNP, the sequences of the two allele-specific forward primers are reported in columns 5 and 6 and the sequence of the common reverse primer is in column 7; the allele present in the resistant parent SRL1 and in the deposited seeds is reported in column 8 and the allele of the usceptible parent (Susc. Parent) is reported in the last column.
- a set of 74 inbred lines derived from different resistant to moderately resistant F2 from the original mapping population was considered for the validation.
- the level of resistance of these 74 inbred lines was assessed in a pathologic test following the same protocol than described in example 2.
- the Acidovorax resistance level is defined as the mean of two separate resistance level measures. Different level of resistance was observed in the set of the 74 inbred lines (see FIG. 4) suggesting the loss of the resistance allele at QTL3 during the fixation process in inbreeds showing low resistance level.
- SNPs A total of 40 additional SNPs have been genotyped on the panel of 74 inbreds.
- the SNPs have been selected within the region of QTL3 defined by the interval between markers CS-0019663 and CS- 0015357 (35 cM) which delimit the physical interval containing the QTL.
- Each single SNP profile has been compared to the resistance level profile of the 74 tested inbred lines.
- the marker profiles explaining the better the resistance differences between the 74 inbreds have been retained to constitute a haplotype.
- the haplotype based on the two markers: CS-0015357 and CS-0022531 explained the most important part of the level of resistance (see FIG. 5).
- CS-0022531 is the best predictive marker for the presence of the QTL conferring the resistance. Its physical position, as well as the physical positions of the most important SNPs on the contig tig00019526 are as follows:
- RR stands for presence (homozygously) of the resistant alleles for both SNPs CS-0015357 and CS-0022531 ,
- RS stands for presence (heterozygously) of the resistant allele and of the susceptible allele for both SNPs CS-0015357 and CS-0022531 ,
- SS stands for presence (homozygously) of the susceptible alleles for both SNPs.
- Seeds of a corn salad varieties are to be treated with EMS by submergence of approximately 2000 seeds per variety into an aerated solution of either 0.5% (w/v) or 0.7% EMS for 24 hours at room temperature.
- M2 seeds are harvested and bulked in one pool per variety per treatment.
- the resulting pools of M2 seeds are used as starting material to identify the individual M2 seeds and the plants with an intermediate resistance against A. valerianellae.
- Rhoads A Au KF (2015). PacBio Sequencing and Its Applications. Genomics proteomics Bioinformatics 13: 278-289. Stam P. (1993). Construction of integrated genetic linkage maps by means of a new computer package : JoinMap. The Plant Journal 3: 739-744.
Landscapes
- Life Sciences & Earth Sciences (AREA)
- Health & Medical Sciences (AREA)
- Physiology (AREA)
- Botany (AREA)
- Developmental Biology & Embryology (AREA)
- Environmental Sciences (AREA)
- Breeding Of Plants And Reproduction By Means Of Culturing (AREA)
Abstract
The invention relates to a corn salad plant with intermediate resistance to Acidovorax valerianellae comprising in its genome a quantitative trait locus (QTL), wherein said QTL confers to the plant resistance to Acidovorax valerianellae and wherein said QTL can be identified with the molecular marker CS-0022531 (SEQ ID NO: 14); said QTL is preferably present in the genome of the seeds of plant ValeriacidoR, NCIMB accession number 43500. The QTL is preferably characterized by defined alleles of several SNPs, especially by a combination of two SNPs forming a haplotype. The invention is also directed to parts of these plants with A. valerianellae resistance phenotype, as well as progeny, to the use of these plants for introgressing the resistance in another genetic background, as well as to different methods for obtaining corn salad plants or seeds with increased resistance to infection by A. valerianellae.
Description
RESISTANCE TO ACIDOVORAX VALERIANELLAE IN CORN SALAD
The present invention relates to resistance, especially intermediate resistance, in corn salad plants, namely plants of Valerianella locusta or Valerianella locusta L, to bacterial spot disease caused by the bacteria Acidovorax valerianellae. According to the invention, the resistance is provided by DNA sequences, or QTL, introgressed into the genome of Valerianella locusta plants. The introgressed sequences can be present homozygously or heterozygously in the genome of a Valerianella locusta L. plant. The present invention also relates to corn salads, comprising the DNA sequences or QTL that lead to resistance to A. valerianellae. The invention further relates to markers linked to said DNA sequences and to the use of such makers to identify or select the DNA sequences and to identify or select plants carrying such resistance. The invention also relates to the seeds and progeny of such plants and to propagation material for obtaining such plants, and to different uses of these plants.
Background of the invention
Corn salad, also known as lamb’s lettuce as well as under its latin name Valerianella locusta L., or Valerianella locusta, is a diploid autogamous crop (2n=14), mostly produced in western part of France (Gardan L. et al., 2003), but which cultivation spreads worldwide. In Europe, main production areas are France and Italy, which together amount for about 85% of world production (about 100 000 tons per year). Production acreage is somewhere near the 18 000-ha including 8000-ha in France, 4600 in Italy and 2700 in Germany, for a seed market of about 110 billion of seed per year.
Corn salad breeding relies mainly on phenotypic selection (Muminovic J. et al. 2004). According to the standard criteria of International Union for the Protection of New Varieties of Plants (UPOV), Corn salad varieties are described and differentiated using morphological traits such as seed size and shape, leaf length, shape, profile, glossiness, color, thickness, and prominence of veins. Genetic studies were limited to amplified fragment length polymorphism (AFLP) fingerprinting to estimate the genetic diversity of corn salad germplasm (Vos P. et al. 1995; Muminovic J. et al. 2004). Conventional breeding aims at producing varieties with round and dark green leaves and is successful in developing such plant cultivars and germplasm. Yet, developing disease resistances is still costly and difficult, especially because of poor genomic resources dedicated to this species.
Bacterial black spot caused by Acidovorax valerianellae (Gardan L. et al., 2003) is a major disease of corn salad in France and is responsible for about 10% losses (in tonnage) on average per year. It was first observed in 1991 , first localized in western France, in high technology field production for the preparation of ready to use salads. It is now spread more globally in all production areas (including Belgium, Germany and Austria). Its occurrence increases with the number of successive crops without break between crop (Grondeau & Samson, 2009). Black and greasy spots on leaves and cotyledons reduce corn-salad quality significantly, and the harvest of infected crops is
unmarketable when the number of spotted plants exceeds 5-10%. The spots could also be framed by a yellow halo and progressively, the whole leaf could become yellow.
In naturally infested fields, soil transmission was demonstrated both indirectly by soil fumigation with metam sodium applied before sowing, which reduced disease incidence and directly by the isolation of the causal organism from soil and root debris up to 39 days after harvest (Grondeau et al., 2007), and by the detection of A. valerianellae in corn-salad seeds, seed transmission of the pathogen and disease development in the field (Grondeau, C. and Samson, R., 2009).
While ground disinfection is less and less efficient, the further decrease of available chemical protection agents results in major problems for cultivation of lamb’s lettuce.
Genetic resistance to A. valerianellae in corn salad would be beneficial in the control of the black spot disease. Such resistance would not only increase the stability of crop protection, but would also result in a reduced, or eliminated, requirement for environmentally harmful, and not always effective, fungicide applications. However, about 30 years after the first identification of the disease, no genetic control of the A. valerianellae is available for V. locusta L. production.
Therefore, there is an important need in the art to identify a reliable source of resistance to A. valerianellae which could be used to obtain resistant commercial cultivated corn salad plants.
Summary of the invention:
The present inventors have identified a resistance against the A. valerianellae pathogen in a wild Valerianella locusta f. carinata (also known as Valerianella carinata) plant and have been able to introgress this resistance into V. locusta L. plants, thus obtaining resistant V. locusta L. plants. The resistance of the present invention is imparted by the newly discovered sequences conferring the resistance, said resistance being linked to an additive quantitative trait loci (QTL), transferable to different V. locusta L genetic backgrounds.
The present invention provides cultivated corn salads that display resistance against the A. valerianellae pathogen as well as methods that produce or identify corn salads that exhibit resistance against A. valerianellae. The present invention also discloses molecular genetic markers, especially Single Nucleotide Polymorphisms (SNPs), linked to the QTL that lead to resistance against the A. valerianellae pathogen. Plants obtained through the methods and uses of such molecular markers are also provided.
Said resistance is moreover easily transferable to different genetic backgrounds and the invention also extends to different methods allowing the transfer or introgression of the QTL conferring the phenotype.
The invention also provides several methods and uses of the information linked to the SNPs associated to the QTL conferring A. valerianellae resistance, inter alia methods for identifying A. valerianellae resistant plants and methods for identifying further molecular markers linked to this resistance to bacterial spot disease caused by A. valerianellae , as well as methods for improving the yield of corn salad production in an environment infected by A. valerianellae and methods for protecting a corn salad culture from A. valerianellae infection or transmission and for controlling the back spot disease caused by A. valerianellae.
Definitions:
The term “Resistance” is as defined by the ISF (International Seed Federation) Vegetable and Ornamental Crops Section for describing the reaction of plants to pests or pathogens, and abiotic stresses for the Vegetable Seed Industry.
Specifically, by resistance, it is meant the ability of a plant variety to restrict the growth and/or development of a specified pest or pathogen and/or the damage it causes when compared to susceptible plant varieties under similar environmental conditions and pest or pathogen pressure. Resistant varieties may exhibit some disease symptoms or damage under heavy pest or pathogen pressure. Two levels of resistance are defined.
High resistance (HR): plant varieties that highly restrict the growth and/or development of the specified pest or pathogen and/or the damage it causes under normal pest pressure when compared to susceptible varieties. These plant varieties may, however, exhibit some symptoms or damage under heavy pest/pathogen pressure.
Intermediate resistance (IR): plant varieties that restrict the growth and/or development of the specified pest or pathogen and/or the damage it causes but may exhibit a greater range of symptoms or damage compared to high resistant varieties. Intermediate resistant plant varieties will still show less severe symptoms or damage than susceptible plant varieties when grown under similar environmental conditions and/or pest pressure.
The disease symptoms of A. valerianellae infection are black and greasy spots on leaves and cotyledons, rendering the salad unmarketable. The spots could also be framed by a yellow halo and progressively, the whole leaf could become yellow.
Susceptibility is the inability of a plant variety to restrict the growth and/or development of a specified pest or pathogen.
A susceptible plant thus displays the detrimental symptoms linked to the bacterial infection, namely the foliar damages.
As used herein, the term “offspring” or “progeny” refers to any plant resulting as progeny from a vegetative or sexual reproduction from one or more parent plants or descendants thereof. For instance, an offspring plant may be obtained by cloning or selfing of a parent plant or by crossing two parental plants and include selfings as well as the F1 or F2 or still further generations. An F1 is a first-generation offspring produced from parents at least one of which is used for the first time as donor of a trait, while offspring of second generation (F2) or subsequent generations (F3, F4, etc.) are specimens produced from selfings of FTs, F2's etc. An F1 may thus be (and usually is) a hybrid resulting from a cross between two true breeding parents (true-breeding is homozygous for a trait), while an F2 may be (and usually is) an offspring resulting from self-pollination of said F1 hybrids.
As used herein, the term “cross”, “crossing”, “cross pollination” or “cross-breeding” refer to the process by which the pollen of one flower on one plant is applied (artificially or naturally) to the ovule (stigma) of a flower on another plant.
As used herein, the term “genetic determinant” and/or “QTL” refers to any segment of DNA associated with a biological function. Thus, QTLs and/or genetic determinants include, but are not limited to, genes, coding sequences and/or the regulatory sequences required for their expression.
QTLs and/or genetic determinants can also include nonexpressed DNA segments that, for example, form recognition sequences for other proteins.
As used herein, the term “genotype” refers to the genetic makeup of an individual cell, cell culture, tissue, organism (e.g., a plant), or group of organisms.
As used herein, the term “heterozygote” refers to a diploid or polyploid individual cell or plant having different alleles (forms of a given gene, genetic determinant or sequences) present at least at one locus.
As used herein, the term “heterozygous” refers to the presence of different alleles (forms of a given gene, genetic determinant or sequences) at a particular locus.
As used herein, the term “homozygote” refers to an individual cell or plant having the same alleles at one or more loci on all homologous chromosomes.
As used herein, the term “homozygous” refers to the presence of identical alleles at one or more loci in homologous chromosomal segments.
As used herein, the term “hybrid” refers to any individual cell, tissue or plant resulting from a cross between parents that differ in one or more genes.
As used herein, the term “locus” (plural: “loci”) refers to any site that has been defined genetically, this can be a single position (nucleotide) or a chromosomal or genomic region. A locus may be a gene, a genetic determinant, or part of a gene, or a DNA sequence, and may be occupied by different sequences. A locus may also be defined by a SNP (Single Nucleotide Polymorphism), by several SNPs, or by two flanking SNPs.
The invention encompasses plants of different ploidy levels, essentially diploid plants, but also triploid plants, tetraploid plants, etc.
In the context of the present invention, DNA strand and allele designation and orientation for the SNP markers is done according to the TOP/BOT method developed by lllumina (https://www.illumina.com/documents/products/technotes/technote_topbot.pdf).
Detailed description of the invention:
The present inventors have identified a QTL which, when present in Valerianella locusta L. plants, provides resistance, essentially intermediate resistance to these plants when infected or likely to be infected by the A. valerianellae bacteria.
The seeds and plants according to the invention have been obtained from an initial cross between a wild plant of Valerianella carinata or Valerianella locusta f. carinata plant, the introgression partner displaying the phenotype of interest, and a plant of Valerianella locusta L, the recurrent susceptible parent, in order to transfer the resistance into Valerianella locusta L. genetic background.
Seeds of resistant Valerianella locusta plants, deriving from this initial cross, which are homozygously resistant against A. valerianellae , and called ValeriacidoR, have been deposited by the inventors at the NCIMB under the accession number NCIMB 43500 on 17th October 2019. The plants grown from these deposited seeds are corn salads resistant against A. valerianellae , namely displaying at least an intermediate resistance to bacterial spot disease caused by A. valerianellae.
According to a first aspect, the present invention is thus directed to a corn salad plant or seed, which is resistant against A. valerianellae , comprising in its genome a quantitative trait locus (QTL), conferring said resistance against. A. valerianellae. Said QTL conferring the resistance was initially introgressed from a wild V. locusta f. carinata, also known as V. carinata plant and are thus referred to as the resistance QTL or introgressed sequences of the invention in the following description. The invention is also directed to a cell of such a plant or seed, comprising these introgressed sequences conferring the resistance.
One would note in this regard that the introgressed resistance QTL has been first identified in a Valerianella locusta f. carinata plant and subsequently transferred into Valerianella locusta L. genetic background. Both Valerianella locusta f. carinata and Valerianella locusta L. are generally recognized as belonging to different species difficult to cross. One man-made interspecific cross has indeed been reported in WO2014009555, but only few interspecific plants were regenerated from it. The identification of a potential resistance to the A. valerianellae pathogen in V. locusta f. carinata background, with a view to introgressing the underlying sequences into V. locusta genetic background was thus a non-obvious breeding program, in view of the reported difficulties.
The resistance phenotype can be tested and scored as described in example 1 of the experimental section, by natural infection or by artificial inoculation for example by vaporization. The resistance against A. valerianellae according to the invention is best characterized by intermediate resistance.
The introgressed sequences conferring the resistance are preferably identified with the SNP (Single Nucleotide Polymorphism) molecular marker CS-0022531 (SEQ ID NO:14). The inventors have indeed demonstrated in the example section that this polymorphism is linked to the resistance QTL and is inherited with the phenotype of interest. This SNP can thus be used as a marker of the presence or absence of the resistance QTL or introgressed sequences. Preferably, the presence of the introgressed sequences, or QTL, conferring the resistance phenotype is identified by allele A of SNP marker CS-0022531.
The specific polymorphisms corresponding to the SNPs referred to in this description, as well as the flanking sequences of these SNPs in the V. locusta genome, are given in the experimental section, in table 2 and in the accompanying sequence listing.
It is to be noted in this respect that, by definition, a SNP refers to a single nucleotide in the genome, which is variable depending on the allele which is present, whereas the flanking nucleotides are identical. For ease of clear identification of the position of the different SNPs, the SNPs are disclosed in table 2 also by reference to theirflanking sequences, identified by SEQ ID number. In the sequence associated with a specific SNP in the present application, for example SEQ ID NO:14 for the SNP CS-0022531 , only one nucleotide within the sequence actually corresponds to the polymorphism, namely the 151st nucleotide of SEQ ID NO:14 corresponds to the polymorphic position of SNP CS- 0022531 , which can be A or G as indicated in table 2. The flanking sequences are given for positioning the SNP in the genome but are not part of the polymorphism as such.
The inventors have moreover shown that the presence of the QTL conferring the resistance
phenotype of the invention can also be tested by detection of a specific haplotype, namely a specific combination of alleles for two SNPs, which combination is informative and representative of the presence of the sequences conferring the resistance. This haplotype is preferably the haplotype corresponding to allele A of SNP CS-0022531 (SEQ ID NO: 14) in combination with allele A of SNP CS-0015357 (SEQ ID NO:9). Consequently, the presence of the QTL conferring the resistance phenotype can be tested or confirmed by the detection of the specific combination of allele A of CS- 0022531 and allele A of CS-0015357.
The QTL according to the invention and conferring the intermediate resistance against A. valerianellae is preferably chosen from the ones present in the genome of seeds of ValeriacidoR. A sample of this Valerianella locusta seed has been deposited by HM. Clause S.A. Rue Louis Saillant, Z.l. La Motte 26800 Portes-les-Valence, France, pursuant to and in satisfaction of the requirements of the Budapest treaty on the International Recognition of the deposit of Microorganisms for the Purpose of Patent procedure (“the Budapest Treaty” with the National collection of Industrial, Food and Marine bacteria (NCIMB) (NCIMB, Ltd, Ferguson Building, Craibstone Estate, Bucksburn, Aberdeen AB21 9YA, united Kingdom) on 17th October 2019 under accession number NCIMB 43500. A deposit of this corns salad seed is maintained by HM. Clause S.A.
The plants grown from these deposited seeds are plants resistant against A. valerianellae infection. A QTL of the invention is thus preferably identical to introgressed sequences from V. carinata present in the genome of the seeds of ValeriacidoR, NCIMB 43500.
The present inventors have also localized the QTL/introgressed sequences responsible for the phenotype of interest. However, insofar as there is no publicly available chromosome map for V. locusta, the QTL has not been localized with respect to the chromosomes of V. locusta, but on the basis of the linkage groups, as identified by the inventors, using classical genotyping approaches and tools described in example 3.
On the basis of the linkage group map elaborated by the inventors, and of the sequencing carried out, the QTL responsible for the phenotype of interest is to be found within a genomic region or interval which comprises or is delimited by SNP CS-0019663 (SEQ ID NO:7) and SNP CS-0021333 (SEQ ID NO: 10), preferably in the region comprising or delimited by SNP CS-0019663 (SEQ ID NO:7) and SNP CS-0015357 (SEQ ID NO:9), and even more preferably in the region comprising or delimited by SNP CS-0016322 (SEQ ID NO:8) and SNP CS-0015357 (SEQ ID NO:9). All these SNP are to be found on Linkage Group 3 (LG3) as identified by the inventors; they are also found on the same contig, and thus are on the same chromosome.
A genomic region identified by flanking sequences, e.g. SNPs, is defined clearly and non- ambiguously.
According to a preferred embodiment, the QTL, or introgressed sequences from V. carinata, are located within a genomic interval delimited by CS-16322 (SEQ ID NO:8) and CS-0015357 (SEQ ID NO:9), or overlaps with said interval.
A genomic region delimited by two SNPs X and Y refers to the section of the genome, more specifically of a chromosome, lying between the positions of these two SNPs and preferably
comprising said SNPs, therefore the nucleotide sequence of this chromosomal region begins with the nucleotide corresponding to SNP X and ends with the nucleotide corresponding to SNP Y, i.e. the SNPs are comprised within the region they delimit, according to the invention.
By “introgressed sequences from V. carinata “ in a given genomic region, it is to be understood that the genomic sequences found in this given region have the same sequence as the corresponding genomic sequences found in the V. carinata donor, i.e. in the introgression partner, at the same locus and also the same sequence as the corresponding genomic sequences found in ValeriacidoR (NCIMB 43500) at the same locus. By having the “same sequence”, it means that the two sequences to be compared are identical to the exception of potential point mutations which may occur during transmission of the genomic region to progeny, i.e. preferably at least 99% identical on a length of 1 kbase. It can be deduced that a genomic region under test has the same sequence, in the sense of the invention, as the corresponding genomic region found in the V. carinata donor at the same locus, if said genomic region under test is also capable of conferring resistance to A. valerianellae.
The presence of introgressed sequences into the genome of a Valerianella locusta plant, seed or cell may for example be shown by GISH (genetic in situ hybridization). GISH is indeed a powerful technique for detection of the introgression of chromatin material from one species or subspecies onto another species or subspecies. The advantage of GISH is that the introgression process is visualized by means of ‘pictures of the introgressed genome’. With this technique, it is also possible to establish if a particular region of the genome is homozygous or heterozygous, thanks to the use of molecular cytogenetic markers which are co-dominant. By this technique, it is also possible to determine in which chromosome or linkage group an introgressed gene of interest is present.
Insofar as the corn salad is autogamous, most of the commercial plants are open pollinated varieties, which are thus essentially homozygous. A corn salad plant, cell or seed of the invention is thus preferably also homozygous for the QTL, or introgressed sequences of the invention conferring A. valerianellae resistance. The invention is however not limited to such homozygous plants, cells or seeds. Indeed, the inventors have also demonstrated that an intermediate resistance is also present in plants having heterozygously the QTL of interest (see experimental section). The present invention thus also encompasses corn salad plant, cell or seed having heterozygously in their genome the QTL, or introgressed sequences, of the invention as defined above.
The inventors have identified and mapped the QTL imparting the intermediate resistance of the invention, mainly by identifying the presence of sequences representative of the introgressed QTL at different loci, namely at least at 5 different loci defined by the 5 following SNPs: CS-0019663 (SEQ ID NO:7), CS-0016322 (SEQ ID NO:8), CS-0015357 (SEQ ID NO:9), CS-0021333 (SEQ ID NO:10) and CS-0022531 (SEQ ID NO:14). The alleles of these molecular markers representative of the QTL or introgressed sequences conferring the resistance of the invention are allele A of CS-0019663, allele A of CS-0016322, allele A of CS-0015357, allele A of CS-0021333 and allele A of CS-0022531. The presence of the QTL in the genome of a corn salad plant, cell or seed according to the invention can thus be detected or revealed by detecting sequences representative of the QTL at said loci, more
preferably by detecting one or more of the following alleles of the corresponding molecular markers: allele A of CS-0019663, allele A of CS-0016322, allele A of CS-0015357, allele A of CS-0021333 and allele A of CS-0022531.
Preferably, the QTL of the invention conferring resistance against A. valerianellae can be detected in a plant, cell, seed, or any other plant part by one or more of the following molecular markers: CS- 0022531 , CS-0015357 and/or CS-0016322. More preferably, said QTL is detected by any one of allele A of CS-0016322, allele A of CS-0015357 and/or allele A of CS-0022531.
Insofar as the QTL confers intermediate resistance when present at the homozygous and heterozygous states, its presence can thus be detected in a plant, cell, seed or any plant part, by detecting one or more of the following genotypes of markers CS-0022531 , CS-0015357 and/or CS- 0016322:
Genotype AA or AG of CS-0022531 ,
Genotype AA or AG of CS-0015357, and/or Genotype AA or AG of CS-0016322.
A plant, seed or cell of the invention, is thus characterized by at least one of the above genotypes, preferably by the genotype AA or AG of CS-0022531 , or by one of the genotypes (AA, AA), (AA, AG), (AG, AA) and (AG, AG) for (CS-0022531 , CS-0015357).
Preferably, a corn salad plant, cell or seed according to the invention is a Valerianella locusta L. plant, a Valerianella locusta L. locusta plant or a Valerianella locusta plant. A corn salad plant, cell or seed according to the invention is not from the wild species Valerianella locusta L. carinata, or Valerianella carinata.
According to a most preferred embodiment, a corn salad plant of the invention is a cultivated plant or line, more preferably an edible, commercial, cultivated corn salad plant. Such a commercial plant or line may also exhibit resistance to other viruses or pests. These preferred embodiments are also applicable to cells, seeds and other plant parts according to the invention.
According to a preferred embodiment, the corn salad plant, cell or seed, or other plant part, of the invention also comprises another type of resistance or intermediate resistance to A. valerianellae , which is not conferred by the introgressed sequences of the invention, but by any other gene or QTL conferring said resistance or intermediate resistance, in order to provide high resistance to the plant, cell or seed, or other plant part, of the invention.
As detailed above, the invention is directed to corn salad plants, exhibiting resistance against bacterial spot disease caused by A. valerianellae bacteria, as well as to seeds giving rise to those plants, and cells of these plants or seeds, or other plant parts, comprising the resistance QTL in their genome, introgressed from V. carinata.
The invention encompasses plants, seeds and cells of any ploidy levels, it encompasses inter alia diploid, triploid, tetraploid and/or allopolyploid plants, cells or seeds.
A plant or seed according to the invention may be a progeny or offspring of a plant grown from the deposited seeds ValeriacidoR, deposited at the NCIMB under the accession number NCIMB 43500. Plants grown from the deposited seeds are indeed homozygous for the QTL of the invention conferring the intermediate resistance of interest, they thus bear in their genome the QTL of interest homozygously. They can be used to transfer these introgressed sequences into another background by crossing and selfing and/or backcrossing.
The invention is also directed to the deposited seeds of ValeriacidoR (NCIMB 43500) and to plants grown from one of these seeds. These seeds contain homozygously the QTL conferring the phenotype of interest.
The invention is also directed to plants or seeds as defined above, i.e. containing the resistance QTL of interest in homozygous or heterozygous state, wherein these plants or seeds are obtainable by transferring the QTL from a Valerianella locusta L. plant, representative seeds thereof were deposited under NCIMB accession NCIMB 43500, into another Valerianella locusta L. genetic background, for example by crossing said deposited plant with a second corn salad parent and selection of the plant or plants bearing the QTL responsible for the phenotype of interest. During such crossing, the QTL can be transferred.
It is noted that the seeds or plants of the invention may also be obtained by different processes not necessarily implying crossing and introgression, and are not exclusively obtained by means of an essentially biological process.
According to such an aspect, the invention relates to a cultivated corn salad plant or seed, preferably a non-naturally occurring corn salad plant or seed, which may comprise one or more mutations in its genome, which provides the plant with resistance against A. valerianellae , which mutation is as present, for example, in the genome of plants of which a representative sample was deposited with the NCIMB under deposit number NCIMB 43500.
In another embodiment, the invention relates to a method for obtaining a corn salad plant or seed carrying one or more mutations in its genome, which provides the plant with resistance against A. valerianellae. Such a method is illustrated in example 6 and may comprise: a) treating M0 seeds of a corn salad to be modified with a mutagenic agent to obtain M1 seeds; b) growing plants from the thus obtained M1 seeds to obtain M1 plants; c) producing M2 seeds by self-fertilization of M1 plants; and d) optionally repeating step b) and c) n times to obtain M1+n seeds.
The M1+n seeds are grown into plants and submitted to A. valerianellae infection. The surviving plants, or those with the milder symptoms of A. valerianellae infection, are multiplied one or more further generations while continuing to be selected for their resistance against A. valerianellae.
In this method, the M1 seeds of step a) can be obtained via chemical mutagenesis such as EMS mutagenesis. Other chemical mutagenic agents include but are not limited to, diethyl sufate (des), ethyleneimine (ei), propane sultone, N-methyl-N-nitrosourethane (mnu), N-nitroso-N-methylurea (NMU), N-ethyl-N-nitrosourea(enu), and sodium azide.
Alternatively, the mutations are induced by means of irradiation, which is for example selected from x-rays, fast neutrons, UV radiation.
In another embodiment of the invention, the mutations are induced by means of genetic engineering. Such mutations also include the integration of sequences conferring the resistance against A. valerianellae , as well as the substitution of residing sequences by alternative sequences conferring the resistance against A. valerianellae. Preferably, the mutations are the integration of the QTL as described above, in replacement of the homologous sequences of a V. locusta L. plant. Even more preferably, the mutations are the substitutions of the sequences comprised within SNP CS-0019663 and SNP CS-0021333 of the Valerianella locusta L. genome, or fragments thereof, by the homologous sequences, present in the genome of a plant of which a representative sample was deposited with the NCIMB under deposit number NCIMB 43500, wherein the sequences or fragments thereof confer resistance against A. valerianellae.
The genetic engineering means which can be used include the use of all such techniques called New Breeding Techniques which are various new technologies developed and/or used to create new characteristics in plants through genetic variation, the aim being targeted mutagenesis, targeted introduction of new genes or gene silencing (RdDM). Example of such new breeding techniques are targeted sequence changes facilitated through the use of Zinc finger nuclease (ZFN) technology (ZFN-1 , ZFN-2 and ZFN-3, see U.S. Pat. No. 9,145,565), Oligonucleotide directed mutagenesis (ODM), Cisgenesis and intragenesis, Grafting (on GM rootstock), Reverse breeding, Agro-infiltration (agro-infiltration "sensu stricto", agro-inoculation, floral dip), Transcription Activator-Like Effector Nucleases (TALENs, see U.S. Pat. Nos. 8,586,363 and 9,181 ,535), the CRISPR/Cas system (see U.S. Pat. Nos. 8,697,359; 8,771 ,945; 8,795,965; 8,865,406; 8,871 ,445; 8,889,356; 8,895,308; 8,906,616; 8,932,814; 8,945,839; 8,993,233; and 8,999,641 ), engineered meganuclease reengineered homing endonucleases, DNA guided genome editing (Gao et al., Nature Biotechnology (2016)), and Synthetic genomics. A major part of targeted genome editing, another designation for New Breeding Techniques, is the applications to induce a DNA double strand break (DSB) at a selected location in the genome where the modification is intended. Directed repair of the DSB allows for targeted genome editing. Such applications can be utilized to generate mutations (e.g., targeted mutations or precise native gene editing) as well as precise insertion of genes (e.g., cisgenes, intragenes, or transgenes). The applications leading to mutations are often identified as site-directed nuclease (SDN) technology, such as SDN1 , SDN2 and SDN3. For SDN1 , the outcome is a targeted, non-specific genetic deletion mutation: the position of the DNA DSB is precisely selected, but the DNA repair by the host cell is random and results in small nucleotide deletions, additions or substitutions. For SDN2, a SDN is used to generate a targeted DSB and a DNA repair template (a short DNA sequence identical to the targeted DSB DNA sequence except for one or a few nucleotide changes) is used to repair the DSB: this results in a targeted and predetermined point mutation in the desired gene of interest. As to the SDN3, the SDN is used along with a DNA repair template that contains new DNA sequence (e.g. gene). The outcome of the technology would be the integration of that DNA sequence into the plant genome. The most likely application illustrating the use of SDN3 would be the insertion of cisgenic, intragenic, or transgenic expression cassettes at a selected
genome location. A complete description of each of these techniques can be found in the report made by the Joint Research Center (JRC) Institute for Prospective Technological Studies of the European Commission in 2011 and titled “New plant breeding techniques - State-of-the-art and prospects for commercial development”.
The resistance, or intermediate resistance, against A. valerianellae corresponds, according to the invention, to a reduction of at least 20% of the symptoms of A. valerianellae infection on leaves and cotyledons, at around 10 to 20 days after infection, with respect to the symptoms on leaves and cotyledons of a plant devoid of said QTL, preferably a reduction of at least 40%, 50%, 60%, 70% or more of said symptoms.
Preferably, the score rate of a plant of the invention is at least 7 in the scale described in example 1 , i.e. 25% or less of symptoms, preferably 12% or less of symptoms, corresponding to a score of 8 or more, and preferably a score of “9” corresponding to a symptomless plant, in the conditions of infection described in example 1.
The invention in another aspect also concerns any plant likely to be obtained from seed or plants of the invention as described above, and also plant parts of such a plant, and most preferably explant, scion, cutting, seed, fruit, root, rootstock, pollen, ovule, embryo, protoplast, leaf, anther, stem, petiole, and any other plant part such as cell, wherein said plant, explant, scion, cutting, seed, fruit, root, rootstock, pollen, ovule, embryo, protoplast, leaf, anther, stem, petiole, and/or plant part is obtainable from a seed or plant according to the first aspect of the invention, i.e. bearing the QTL of interest, also called introgressed sequences of the invention, in their genome. These plant parts, inter alia explant, scion, cutting, seed, fruit, root, rootstock, pollen, ovule, embryo, protoplast, leaf, anther, stem or petiole, comprise in their genome the QTL conferring the phenotype of interest, i.e. resistance against A. valerianellae ; they thus comprise a cell comprising this resistance QTL or introgressed sequences from V. carinata. As mentioned in respect of the first aspect of the invention, this QTL or introgressed sequences is/are preferably as found in the genome of the deposited seed ValeriacidoR NCIMB 43500.
According to a preferred embodiment, the invention is directed to seed as described above, which develops into a plant according to the first aspect of the invention, thus resistant against A. valerianellae infection thanks to the presence of the resistance QTL, or introgressed sequences, as defined above.
The invention is also directed to cells of V. locusta plants, such that these cells comprise, in their genome, the introgressed QTL of the present invention conferring resistance against A. valerianellae to a V. locusta plant. The QTL is as already defined in the context of the present invention, it is characterized by the same features and preferred embodiments already disclosed with respect to the plants and seeds according to the preceding aspects of the invention. The presence of this QTL, or introgressed sequences, can be revealed by the techniques disclosed above and well known to the skilled reader. The QTL can inter alia be identified in the genome of such a cell of the invention
with the molecular marker CS-0022531 , especially allele A of this marker. It is advantageously characterized by the presence of the haplotype corresponding to allele A of CS-0022531 and allele A of CS-0015357. Said QTL can be present homozygously or heterozygously.
Cells according to the invention can be any type of V. locusta L. or V. locusta cell, inter alia an isolated cell and/or a cell capable of regenerating a whole corn salad, bearing the QTL or introgressed sequences of interest. The cell is not a cell of the wild species V. carinata.
The present invention is also directed to a tissue culture of non-regenerable or regenerable cells of the plant as defined above according to the present invention; preferably, the regenerable or non- regenerable cells are derived from embryos, protoplasts, meristematic cells, callus, pollen, leaves, anthers, stems, petioles, roots, root tips, fruits, seeds, flowers, cotyledons, and/or hypocotyls of the invention, and the cells contain the QTL, or introgressed sequences, of interest, homozygously or heterozygously in their genome conferring resistance against A. valerianellae.
The tissue culture will preferably be capable of regenerating plants having the physiological and morphological characteristics of the foregoing corn salad, and of regenerating plants having substantially the same genotype as the foregoing corn salad plant. The present invention also provides corn salad plants regenerated from the tissue cultures of the invention.
The invention also provides a protoplast of the plant defined above, or from the tissue culture defined above, said protoplast containing the QTL, or introgressed sequences, conferring the resistance phenotype of the invention.
The invention is also directed to tissue of a plant of the invention; the tissue can be an undifferentiated tissue, or a differentiated tissue. Such a tissue comprises one or more cells comprising the QTL of the invention.
The invention is also directed to propagation material, capable of producing a resistant corn salad according to the invention, comprising the introgressed sequences or QTL as defined above.
The invention also provides food product comprising the corn salad of the invention, or parts thereof, especially leaves or young shoots, which comprise cells having the introgressed QTL of the invention in their genome.
According to another aspect, the present invention is also directed to the use of a corn salad plant of the invention, preferably comprising homozygously the QTL of the invention, as a breeding partner in a breeding program for obtaining V. locusta plants, preferably edible corn salad plants having resistance against A. valerianellae , namely resistance against bacterial spot disease caused by A. valerianellae. Indeed, such a breeding partner harbors, preferably homozygously in its genome the QTL, or introgressed sequences, conferring the resistance of interest. By crossing this plant with a corn salad, especially a line or an open pollinated variety, it is thus possible to transfer the QTL of the present invention conferring the desired phenotype, to the progeny. A plant according to the invention, or any part thereof comprising the QTL of interest, can thus be used as a breeding partner for introgressing the QTL, or introgressed sequences, conferring the desired resistance into an edible corn salad or V. locusta L. germplasm and/or for breeding V. locusta plants resistant against A. valerianellae infection.
Although a plant or seed bearing the QTL of interest heterozygously, can also be used as a breeding partner, the segregation of the QTL, or introgressed sequences, is likely to render the breeding program more complex.
The improved phenotype of the invention is resistance against A. valerianellae , namely resistance against bacterial spot disease caused by A. valerianellae. It is preferably to be transferred into a plant which is initially susceptible to A. valerianellae infection. As already mentioned above, it can also be transferred into a plant comprising another form of A. valerianellae resistance, conferred by another QTL, gene or introgression.
The introgressed QTL will advantageously be introduced into varieties or OP varieties that contain other desirable genetic traits such as resistance to others diseases or pest, good agronomical features, drought tolerance, and the like.
The invention is also directed to the same use with plants or seed of ValeriacidoR, deposited at the NCIMB under the accession number NCIMB 43500. Said plants are also suitable as introgression partners in a breeding program aiming at conferring the desired phenotype to V. locusta plant or germplasm.
In such a breeding program, the selection of the progeny displaying the desired phenotype, or bearing the QTL linked to resistance against A. valerianellae can advantageously be carried out on the basis of the alleles of the SNP markers, especially the SNP markers of the invention, and preferably SNP markers CS-0022531 , CS-0015357 and/or CS-0016322, more preferably the alleles of the haplotype constituted by CS-0022531 and CS-0015357, and even more preferably by CS- 0022531.
A progeny is preferably selected on the presence of at least one of the following alleles: allele A of CS-0019663, allele A of CS-0016322, allele A of CS-0015357, allele A of CS-0021333 and allele A of CS-0022531. The progeny is advantageously selected on the basis of the presence of allele A of CS-0022531 , or the combination of allele A of CS-0022531 and allele A of CS-0015357.
The progeny is preferably selected on the presence of one of these alleles homozygously.
Such selection will be made on the presence of the allele or alleles of interest in a genetic material sample of the plant to be selected. The presence of this or these allele(s) indeed confirms the presence of the QTL, or introgressed sequences, of the invention at the loci defined by said SNPs, as found in the deposited seeds.
A plant according to the invention, or grown from a seed as deposited under accession number NCIMB 43500, is thus particularly valuable in a marker assisted selection for obtaining commercial corn salad lines and varieties, having the improved phenotype of the invention, namely resistance against A. valerianellae , more specifically intermediate resistance.
The invention is also directed to the use of said plants in a program aiming at identifying, sequencing and / or cloning the genetic introgressed sequences conferring the desired phenotype.
Any specific embodiment described for the previous aspects of the invention is also applicable to this aspect of the invention, especially with regard to the features of the QTL conferring the phenotype of interest.
According to still another aspect, the invention also concerns methods or processes for the production or breeding of V. locusta plants, especially edible cultivated corn salads, having the desired phenotype, especially commercial plants and inbred parental lines. The present invention is indeed also directed to transferring the QTL, or introgressed sequences of the invention conferring the resistance against A. valerianellae , to other corn salads, especially other corn salad varieties, or other species or inbred parental lines, and is useful for producing new types and varieties of corn salad.
In this regard, the invention also comprises methods for breeding V. locusta plants having resistance against A. valerianellae , comprising the steps of crossing a plant grown from the deposited seeds ValeriacidoR NCIMB 43500 or progeny thereof bearing the QTL of the invention conferring A. valerianellae resistance, with an initial V. locusta plant preferably devoid of said QTL. The QTL is as defined above, namely introgressed from V. carinata and is preferably present in the genome of the seeds of ValeriacidoR, NCIMB accession number 43500. This QTL is identifiable by CS-0022531 , preferably by allele A of said SNP, and also preferably by the haplotype corresponding to allele A of CS-0022531 and allele A of CS-0015357.
The invention also concerns a method for conferring resistance against A. valerianellae to a V. locusta plant, comprising genetically modifying said plant to introduce the resistance QTL of the invention. Said QTL is as defined above and is preferably present in the genome of the seeds of ValeriacidoR, NCIMB accession number 43500. The genetic modification can be carried out by any methods or means well known to the skilled person.
The invention is also directed to a method or process for the production of a plant having resistance against A. valerianellae; such a method or process may comprise the following steps: a) Crossing a plant grown from a deposited seed NCIMB 43500, or progeny thereof comprising the QTL conferring resistance against A. valerianellae , and an initial V. locusta plant, preferably devoid of said QTL, b) Selecting one plant in the progeny thus obtained, bearing the QTL of the present invention; c) Optionally self-pollinating one or several times the plant obtained at step b) and selecting in the progeny thus obtained a plant having A. valerianellae resistance, especially intermediate resistance.
Alternatively, the method or process may comprise instead of step a) the following steps: a1) Crossing a plant corresponding to the deposited seeds (NCIMB 43500), or progeny thereof, bearing the QTL conferring A. valerianellae resistance, and an initial V. locusta plant, preferably devoid of said QTL, a2) Increasing the F1 hybrid by means of selfing to create F2 population.
In the above methods or processes, SNP markers are preferably used in steps b) and / or c), for selecting plants bearing sequences conferring the resistance phenotype of interest.
The SNP markers are preferably one or more of the 5 preferred SNP markers of the invention, namely CS-0019663, CS-0016322, CS-0015357, CS-0021333 and CS-0022531 , including all combinations thereof, especially any combination comprising SNP CS-0022531 , preferably comprising CS-
0015357 and CS-0022531 , inter alia the combination of the three markers CS-0016322, CS-0015357 and CS-0022531.
By selecting a plant on the basis of the allele of one or more markers or SNPs, it is to be understood that the plant is selected as having A. valerianellae resistance, intermediate resistance or improved resistance with respect to the initial plant, when the allele of the SNP(s) is (are) the allele corresponding to the allele of the resistant parent for this SNP and not the allele of the initial susceptible V. locusta plant. Those alleles for the SNPs of the invention have already been detailed in this application and are reported in table 2.
For example, a plant can be selected as having the improved phenotype of the invention, when allele A of CS-0019663, allele A of CS-0016322, allele A of CS-0015357, allele A of CS-0021333 and/or allele A of CS-0022531 is detected.
Preferably, the V. locusta plant of step a) is an elite line, used in order to obtain a plant with commercially desired traits or desired horticultural traits.
A method or process as defined above may advantageously comprise backcrossing steps, preferably after step c), in order to obtain plants having all the characterizing features of V. locusta plants, especially of edible corn salad. Consequently, a method or process for the production of a plant having these features may also comprise the following additional steps: d) Backcrossing the resistant plant selected in step b) or c) with a V. locusta plant; e) Selecting a plant bearing the QTL, or introgressed sequences, of the present invention.
The plant used in step a), namely the plant corresponding to the deposited seeds can be a plant grown from the deposited seeds; it may alternatively be any plant according to the 1st aspect of the invention, bearing the QTL or introgressed sequences conferring the phenotype.
At step e), SNP markers can be used for selecting plants having an improved resistance to A. valerianellae , with respect to the initial plant. The SNP markers are those of the invention, as described in the previous sections.
According to a preferred embodiment, the method or process of the invention is carried out such that, for at least one of the selection steps, namely b), c) and/or e), the selection is based on the detection of at least one of the resistance alleles corresponding to allele A of CS-0019663, allele A of CS- 0016322, allele A of CS-0015357, allele A of CS-0021333 and allele A of CS-0022531 , preferably detection of allele A of CS-0022531 or detection of allele A of CS-0015357 and allele A of CS- 0022531. The preferred alleles and combinations have already been disclosed and are applicable to this embodiment.
The presence of the preferred alleles or combinations is informative regarding the presence of the QTL of the invention, conferring the phenotype of interest.
It is to be noted that, when plants having the improved phenotype, and bearing homozygously the QTL, or introgressed sequences, of the invention, are to be selected, the selection is to be made advantageously on the basis of one or more the SNPs of the invention, on the presence of the alleles representative of the QTL, namely the alleles found in the resistant parent, in combination with the absence of the alleles representative of the recurrent susceptible V. locusta parent.
The selection can also be made on the basis of any other marker linked to the introgressed
sequences and representative of the presence of these introgressed sequences by opposition to the resident sequences of the susceptible parent. Methods for defining alternative markers are also in the scope of the present invention.
The plant selected at step e) is preferably a cultivated or commercial corn salad plant.
Preferably, steps d) and e) are repeated at least twice and preferably three times, not necessarily with the same V. locusta plant. Said V. locusta plant(s) is (are) preferably a breeding line (s).
The self-pollination and backcrossing steps may be carried out in any order and can be intercalated, for example a backcross can be carried out before and after one or several self-pollinations, and self- pollinations can be envisaged before and after one or several backcrosses.
The method used for allele detection can be based on any technique allowing the distinction between two different alleles of a SNP or other marker, at a defined genomic locus.
The present invention also concerns a plant obtained or obtainable by such a method. Such a plant is indeed a V. locusta plant having the resistance phenotype according to the first aspect of the invention.
The invention is also directed to a method for obtaining cultivated or commercial corn salad plants or inbred parental lines thereof, having the desired improved phenotype, corresponding to an improved resistance to A. valerianellae with respect to an initial commercial V. locusta plant, comprising the steps of: a) Backcrossing a plant obtained by germinating a deposited seed ValeriacidoR NCIMB accession number 43500, or progeny thereof, bearing the QTL conferring A. valerianellae resistance, with a commercial corn salad plant, b) Selecting a plant bearing the QTL.
Preferably, the selection is made on the basis of one or more of the 5 SNPs of the invention, as detailed for the other methods of the invention, preferably a selection comprising at least selection of allele A of CS-0022531 , and preferably selection of allele A of CS-0015357 and allele A of CS- 0022531. Alternative marker of the QTL can also be used.
The present invention is also directed to a V. locusta plant and seed obtainable by any of the methods and processes disclosed above. The seed of such V. locusta are preferably coated or pelleted with individual or combined active species such as plant nutrients, enhancing microorganisms, or products for disinfecting the environment of the seeds and plants. Such species and chemicals may be a product that promotes the growth of plants, for example hormones, or that increases their resistance to environmental stresses, for example defense stimulators, or that stabilizes the pH of the substrate and its immediate surroundings, or alternatively a nutrient.
They may also be a product for protecting against agents that are unfavorable toward the growth of young plants, including herein viruses and pathogenic microorganisms, for example a fungicidal, bactericidal, hematicidal, insecticidal or herbicidal product, which acts by contact, ingestion or gaseous diffusion; it is, for example, any suitable essential oil, for example extract of thyme. All these products reinforce the resistance reactions of the plant, and/or disinfect or regulate the environment
of said plant. They may also be a live biological material, for example a nonpathogenic microorganism, for example at least one fungus, or a bacterium, or a virus, if necessary with a medium ensuring its viability; and this microorganism, for example of the pseudomonas, bacillus, trichoderma, clonostachys, fusarium, rhizoctonia, etc. type stimulates the growth of the plant, or protects it against pathogens.
In all the previous methods and processes, the identification of the plants bearing the QTL, or introgressed sequences, responsible for the improved resistance to A. valerianellae could be done by the detection of at least one of the alleles of the SNPs associated with the introgressed resistance QTL, potentially in combination with the absence of the other allelic form of the SNPs of the present invention, in order to confirm the homozygous state of the QTL if needed. As such, the identification of a plant bearing homozygously the QTL or introgressed sequences of the present invention will be based on the identification of at least one of allele A of CS-0019663, allele A of CS-0016322, allele A of CS-0015357, allele A of CS-0021333 and allele A of CS-0022531 , as well as the absence of the other allele for these SNPs, which are as disclosed in table 2.
The invention is also directed to the use of the information provided herewith by the present inventors, namely the existence of a QTL, or introgressed sequences, present in the deposited seeds of ValeriacidoR (NCIMB accession number 43500), and conferring the resistance phenotype to V. locusta plants, and the disclosure of molecular markers associated to this QTL or introgressed sequences. This knowledge can be used inter alia for precisely mapping the QTL, for defining its sequence, for identifying corn salad plants comprising the QTL conferring the resistance phenotype and for identifying further or alternative markers associated to this QTL. Such further markers are characterized by their location, namely close to the 5 markers disclosed in the present invention, preferably close to at least one of the 2 markers of the haplotype, and by their association with the phenotype of interest, as revealed by the invention, namely A. valerianellae resistance, more specifically intermediate resistance against this pathogen.
The invention is thus also directed to a method for identifying, detecting and/or selecting V. locusta plants having the QTL of the present invention as found in the genome of the seeds of ValeriacidoR (NCIMB accession number 43500), said QTL conferring an improved A. valerianellae resistance with respect to a corresponding plant devoid of said QTL, the method comprising the detection of at least one of the following markers: allele A of CS-0019663, allele A of CS-0016322, allele A of CS- 0015357, allele A of CS-0021333 and allele A of CS-0022531 , preferably allele A of CS-0022531 potentially in combination with allele A of CS-0015357, in a genetic material sample of the plant to be identified and/or selected.
The invention is also directed to a method for detecting or selecting V. locusta plants having the QTL conferring A. valerianellae resistance and having at least one of the alleles mentioned above, wherein the detection or selection is made on condition of A. valerianellae infection comprising inoculation of A. valerianellae on the plants to be tested, for example inoculation by vaporization. The presence of the phenotype of interest is informative of the presence of the QTL or introgressed
sequences of the invention.
The invention is also directed to a method for detecting and/or selecting a V. locusta plant, especially cultivated edible corn salad, having the QTL of the invention, comprising the detection of at least one of the alleles mentioned above.
The method is particularly adapted in a breeding program with ValeriacidoR (NCIMB accession number 43500), as initial parent, or progeny thereof, comprising the QTL of the invention conferring A. valerianellae resistance.
The invention is further directed to a method for detecting and/or selecting V. locusta plants, especially cultivated edible corn salad, having the QTL of the present invention conferring the resistance phenotype, on the basis of the detection of any molecular marker revealing the presence of said QTL or informative about the presence of said QTL. Indeed, now that the QTL of the present invention has been identified by the inventors, the identification and then the use of alternative molecular markers, in addition to the SNPs already identified and disclosed by the inventors, can be done without difficulty by the skilled artisan. The QTL may be characterized by the presence of at least one of the SNPs of the invention, especially CS-0022531 , or by the haplotype corresponding to CS-0022531 and CS-0015357, but it may also be identified through the use of different, alternative markers. Also included in the present invention are thus methods and uses of any such molecular markers for identifying the QTL of the invention in a corn salad genome, wherein said QTL confers an improved A. valerianellae resistance with respect to a corresponding plant devoid of said QTL, the QTL being characterized by the presence of at least one of allele A of CS-0019663, allele A of CS-0016322 , allele A of CS-0015357 , allele A of CS-0021333 and allele A of CS-0022531 , preferably at least by allele A of CS-0022531.
Are also included methods and uses of any such alternative molecular marker for identifying the QTL or introgressed sequences of the invention in a corn salad genome, wherein said QTL confers A. valerianellae resistance and is characterized by the presence of at least one of allele A of CS- 0019663, allele A of CS-0016322, allele A of CS-0015357, allele A of CS-0021333 and allele A of CS-0022531 , preferably at least by allele A of CS-0022531.
The invention also concerns a method for detecting and/or selecting corn salad plants having the resistance QTL as defined previously, conferring A. valerianellae resistance, said method comprising: a) Assaying corn salad plants for the presence of at least one genetic marker genetically linked or associated to the QTL involved in A. valerianellae resistance, b) Selecting a plant comprising the genetic marker and the linked or associated QTL involved in A. valerianellae resistance, wherein the QTL and the genetic marker are to be found in the genomic region delimited by CS- 0019663 and CS-0021333 in the genome of V. locusta.
By association, or genetic association, and more specifically genetic linkage, it is to be understood that a genetic polymorphism of the marker (e.g. a specific allele of the SNP marker) and the
phenotype of interest occur simultaneously, i.e. are inherited together, more often than would be expected by chance occurrence, i.e. there is a non-random association of the allele and of the genetic sequences responsible for the phenotype, as a result of their genomic proximity.
A molecular marker of the invention, either one of 5 markers disclosed above or alternative markers, are inherited with the phenotype of interest in preferably more than 90% of the meioses, preferably in more than 95%, 96%, 98% or 99% of the meioses.
The definition and preferred features of the QTL, or introgressed sequences, of the invention are as defined in other sections of the present specification.
The invention thus concerns the use of one or more molecular markers, for fine-mapping or identifying a QTL in the corn salad genome, said QTL participating to the improved phenotype of the invention, wherein said one or more markers is/are localized in one of the following genomic regions:
- in the genomic region delimited by CS-0019663 and CS-0021333,
- at less than 2 megabase units from the locus of one of CS-0016322, CS-0022531 and CS- 0015357.
According to a preferred embodiment, said one or more markers is in the genomic region delimited by CS-0016322 and CS-0021333, preferably by CS-0016322 and CS-0015357, more preferably by CS-0022531 and CS-0015357.
Said one or more molecular marker(s) is/are moreover preferably associated, with a p-value of 0.05 or less, with at least one of the following SNP alleles: allele A of CS-0019663, allele A of CS-0016322, allele A of CS-0015357, allele A of CS-0021333 and allele A of CS-0022531.
The molecular marker is preferably a SNP marker. It is more preferably at less than 1 megabase from the locus of at least one of the 5 SNPs of the invention, preferably of one of the 3 markers CS- 0016322, CS-0015357 and CS-0022531 , and preferably of CS-0022531.
The QTL is to be found in the deposited seeds NCIMB 43500.
The p-value is preferably less than 0.01.
The invention is also directed to the use of at least one of the 5 SNPs of the invention, associated with the QTL conferring A. valerianellae resistance to corn salad, for identifying one or more alternative molecular markers associated with said QTL, wherein said one or more alternative molecular markers are:
- in the genomic region delimited by CS-0019663 and CS-0021333,
- at less than 2 megabase units from the locus of one of CS-0016322, CS-0022531 and CS- 0015357.
According to a preferred embodiment, said one or more markers are in the genomic region delimited by CS-0016322 and CS-0021333, preferably by CS-0016322 and CS-0015357, more preferably by CS-0022531 and CS-0015357. The genetic association or linkage can advantageously be detected by following the alternative maker and the presence of the QTL in the progeny arising from a plant comprising the QTL of interest.
The alternative molecular markers are preferably associated with said QTL with a p-value of 0.05 or less, preferably less than 0.01. The QTL is to be found in the deposited seeds NCIMB 43500.
The QTL mentioned above, conferring A. valerianellae resistance according to the invention, is the introgressed QTL present in ValeriacidoR (NCIMB accession number 43500).
Genetic association or linkage is as defined above; preferably the association or linkage is with a p- value of preferably less than 0.05, and most preferably less than 0.01 or even less.
A molecular marker and the resistance phenotype are inherited together in preferably more than 90% of the meioses, preferably more than 95%.
The molecular markers according to this aspect of the invention are most preferably SNP. They are more preferably at less than 1 megabase from the locus of at least one of the 5 SNPs of the invention, preferably at less than 0.5 megabases.
The invention also comprises a method for identifying a molecular marker associated with a QTL participating to A. valerianellae resistance in corn salad, as described in the present application, comprising the steps of:
- identifying a molecular marker in the genomic region delimited by CS-0019663 and CS- 0021333, or at less than 2 megabase units from the locus of one of CS-0016322, CS-0022531 and CS-0015357, preferably less than 1 or less than 0.5 megabase units; and
- determining whether an allele or state of said molecular marker is associated with the phenotype of A. valerianellae resistance in a segregating population issued from a plant exhibiting A. valerianellae resistance, for example in a segregating population issued from a plant corresponding to the deposited seeds.
The invention is also directed to the use of a molecular marker for identifying or selecting a corn salad plant comprising, in its genome, a QTL conferring to V. locusta plants A. valerianellae resistance, wherein said marker is in the genomic region delimited by CS-0019663 and CS-0021333, or at less than 2 megabase unit from the locus of at least one of the 5 SNP markers of the invention; and wherein said molecular marker is associated with at least one of the following SNP alleles: allele A of CS-0019663, allele A of CS-0016322, allele A of CS-0015357, allele A of CS-0021333 and allele A of CS-0022531 , with a p-value of 0.05 or less, preferably 0.01 or less.
The molecular marker to be used according to this embodiment is obtainable inter alia by the method for identifying further or alternative molecular markers, as disclosed above. The molecular marker is preferably a SNP marker. It is more preferably at less than 1 megabase from the locus of at least one of the 5 SNPs of the invention.
According to still another aspect, the invention is also directed to a method for genotyping a plant, preferably a V. locusta plant or corn salad germplasm, for the presence of at least one genetic marker associated with A. valerianellae resistance, wherein the method comprises the determination or detection in the genome of the tested plant of a nucleic acid comprising at least one of the markers of the invention, or comprising at least one of the alternative molecular markers as disclosed above.
Preferably, the method comprises the step of identifying in a sample of the plant to be tested specific sequences associated with A. valerianellae resistance, in nucleic acids comprising at least one of the alleles of the SNPs of the invention, as already disclosed.
The detection of a specific allele of a SNP can be carried out by any method well known to the skilled reader.
In view of the ability of the resistant plants of the invention to restrict the damages caused by A. valerianellae infection, namely to restrict bacterial spot disease caused by A. valerianellae bacteria, they are advantageously grown in an environment infected or likely to be infected by A. valerianellae ; in these conditions, the resistant plants of the invention produce more marketable corn salad than susceptible plants. The invention is thus also directed to a method for improving the yield of corn salad in an environment infected by A. valerianellae comprising growing corn salads comprising in their genome a QTL, or introgressed sequences, as defined according to the previous aspects of the invention, and conferring to said plants A. valerianellae resistance.
Preferably, the method comprises a first step of choosing or selecting a corn salad plant having said QTL, or introgressed sequences, of interest. The method can also be defined as a method of increasing the productivity of a corn salad field, tunnel, greenhouse or glasshouse, or as a method of reducing the intensity or number of fungicide application in the production of corn salad.
The invention is also directed to a method for reducing the loss on corn salad production in conditions of A. valerianellae infection, especially loss due to unmarketable corn salads, comprising growing a corn salad plant as defined above.
These methods are particularly valuable for a population of corn salad plants, either in a field, in tunnels, greenhouses or in glasshouses.
Alternatively, said methods for improving the yield or reducing the loss on corn salad production may comprise a first step of identifying corn salad plants resistant against A. valerianellae resistance and comprising in their genome a QTL that confers to said plants A. valerianellae resistance, and then growing said resistant plants in an environment infected or likely to be infected by the bacteria.
The resistant plants of the invention are also able to restrict at least partially, the multiplication of A. valerianellae , thus limiting the infection of further plants and the propagation of the bacteria. Accordingly, the invention is also directed to a method of protecting a field, tunnel, greenhouse or glasshouse, or any other type of plantation, from A. valerianellae infection, or of at least limiting the level of infection by A. valerianellae of said field, tunnel, greenhouse or glasshouse or of limiting the spread of A. valerianellae in a field, tunnel, greenhouse or glasshouse, especially in glasshouse. Such a method preferably comprises the step of growing a resistant plant of the invention, i.e. a plant comprising in its genome a QTL conferring to said plant A. valerianellae resistance.
The invention also concerns the use of a plant resistant to A. valerianellae for controlling A. valerianellae infection, or controlling the black spot disease caused by A. valerianellae bacteria, in a field, tunnel, greenhouse or glasshouse, or other plantation; such a plant is a plant of the invention, comprising in its genome a QTL as defined above. According to this use, the plants of the invention
are therefore used for protecting a field, tunnel, greenhouse or glasshouse from A. valerianellae infection.
All the preferred features of the QTL are as defined in connection with the other aspects of the invention, namely it is present in the seeds of ValeriacidoR (NCIMB accession number 43500), and it is identifiable by the SNP marker CS-0022531 , preferably by allele A of CS-0022531 , or identifiable by the haplotype corresponding to allele A of CS-0022531 and allele A of CS-0015357.
LEGEND OF FIGURES:
FIG. 1: Distribution of the Acidovorax score in an F2 population (152 different plants) of SRLIxSusceptible Parent; the X-axis represents the Acidovorax score according to the rating disclosed in example 1. The Y-axis represent the number of progenies.
FIG.2: Genomic positions of the QTLs detected on the Linkage Group 3 (LG 3) for F2 population of -SRL1 x Susceptible”
FIG.3: Genomic position of the QTL detected on the Linkage Group 3 (LG 3) of ‘“SRL1 x Susceptible” map by multiple QTL mapping (MQM) for the Acidovorax resistance score noted on leaves.
X-axis represents the genomic positions of the SNPs along the linkage group 3. The Y-axis represent the LOD Score.
FIG.4: Distribution of Acidovorax scores obtained within the panel of the 74 inbred lines tested for validation; the scores illustrated are the mean of two different measures.
FIG.5: Boxplot of adjusted values of resistance to Acidovorax in the 74 inbred lines, as a function of the genotypes at QTL3 haplotype consisting of SNP markers CS-0015357 and CS-0022531 (RR: Resistant alleles (homozygously) for both SNPs, RS: Heterozygous state for both SNPs, i.e. presence of the Resistant allele and of the susceptible allele for CS-0015357 and CS-0022531 , SS: Susceptible alleles (homozygously) for both SNPs).
FIG.6: Results of analyses of variance of QTL3 on Acidovorax level of resistance. The meaning of RR, RS and SS is as defined for FIG. 5.
EXAMPLES:
Example 1: Pathology test
A French strain of Acidovorax valerianellae has been collected in Nantes area. The inoculum is prepared by multiplying the bacteria on solid King B medium for 48 hours at 26°C and further diluted at 108 bacteria per ml in NaCI 0.9% solution (D.O. 0.3 at 600 nm). Plantlets to be tested are grown in greenhouses for 3 weeks (3-4 leaves old plantlets stage). They are cultivated at 20-24°C in batches of 25 seeds of the genotype to be tested by repetition (total 6 reps). Plantlets are inoculated by vaporization and grown in growth chambers at 21 °C.
For plant rescue tests, seeds are sown on 140 well-trays and grown in net houses until 3-4 leaf stages. After inoculation, they are grown in growth chambers at 21 °C.
The pathology test is read 12 to 14 days after inoculation with a 1 to 9 scale notation for symptoms on leaves and cotyledons, where
“1” correspond to 76% to 100% of symptoms,
“3” correspond to 51% to 75% of symptoms,
“5” correspond to 26% to 50% of symptoms,
“7” correspond to 13% to 25% of symptoms,
“8” correspond to 1% to 12% of symptoms and where “9” corresponds to symptomless plants.
Example 2: Identification of a resistant plant.
Among their internal gene bank composed of 288 accessions, the present inventors identified a wild Valerianella locusta F. carinata (SRL1) showing some intermediate resistance to Acidovorax valerianellae in pathology test set up according to the protocol described in example 1. This accession was further tested five times and always scored among the best accessions, displaying a strong Acidovorax score according to the test described in Example 1.
The SRL1 resistant plant was crossed with a variety of Valerianella locusta L. susceptible to Acidovorax valerianellae, and the F1 was further selfed, thereby creating an F2 population of 152 different plants to be evaluated according to the pathology test of example 1. In addition to the randomized F2 population plants, susceptible and resistant seeds of the parents were planted as controls.
The distribution of the Acidovorax score is represented in FIG. 1 for the studied progenies; the average score for the studied progenies and its parental lines is reported in table 1.
The normal distribution (see Fig. 1) tends to indicate that the resistance behaves as a complex trait relying on a polygenic genetic architecture, rather than a single gene model.
Example 3: Sequencing, genetic analysis and map construction
Due to a lack of genomic resources, neither a genetic map nor detailed marker information are publicly available for Valerianella locusta. As such, the present inventors had to develop a reference genome and specific markers in a three steps process.
In a first step, the variety GALA (HM. Clause) was fully sequenced and assembled in order to obtain a reference sequence ( de novo whole genome sequencing approach - WGS). In a second step, 48 other, different varieties were used in a genotyping by sequencing approach (GBS) in order to identify allelic variations from the GALA reference sequence established in 1. In the last step, 48 F2 plants
from the population established in example 2 were analyzed in order to identify the specific variations existing between the parental material of the said population, especially the ones linked to the Acidovorax valerianellae resistance.
The genotyping by sequencing approach was performed according to the protocol described in Elshire RJ et al., 2011 for the libraries preparation, followed by a next generation lllumina HiSeq sequencing for the genotyping by sequencing itself.
The whole genome sequencing assembly was done using PacBio long read technology (Rhoads A, Au KF, 2015).
Genome-wide SNP calling from GBS data has been performed using TASSEL 5.0 pipeline (Glaubitz JC, et al. 2014).
The Map construction was carried out using JoinMap version 4.0 (JoinMap 4, Software for the calculation of genetic linkage maps in experimental populations. Kyasma BV, Wageningen, Netherlands). Chi square values were calculated for each marker to detect segregation deviations (P<0.05) from the expected Mendelian ratio and highly distorted markers were discarded.
Linkage groups (LG) were determined by a regression mapping procedure using an independence test of the logarithm of the odds (LOD score) with a minimum threshold of 5.0 (Stam P. 1993).
Map construction was performed using the Kosambi mapping function (Kosambi DD; 1944) and the regression mapping algorithm following the JoinMap parameters: Rec=0.40, LOD=1.0, Jump=5.
A total of nine LGs has been identified and drawn using the MapChart software (Voorrips RE, 2002).
Example 4: Quantitative Trait Loci (QTL) Detection
QTL analyses were carried out using Map QTL version 6.0 on the F2 population (Van Ooijen JW 2009).
A first step was the Kruskal-Wallis analysis in which, the effect of the marker significance level on a specific trait, here the Acidovorax valerianellae resistance was estimated by simple linear regression between the marker and the trait (Kenis K, Keulemans J, 2007).
Then, QTL mapping was performed using interval mapping (IM) which estimates the likelihood score of a putative QTL placed in any position within an interval flanked by two adjacent markers (Jansen RC and Stam P, 1994).
The LOD score threshold (P-value of 0.05) at which a QTL was declared significant, was determined using a permutation test for each LG as well as for the genome wide. Over 1000 permutations the frequency distribution of the maximum LOD score under the null hypothesis (no QTL) was determined (Churchill GA, Doerge RW 1994).
When several markers displayed a LOD score equals or superior to the LOD score determined by the permutation test, they were declared as cofactors for a multiple QTL mapping (MQM) analysis with a step size of 1 cM (Jansen RC and Stamp P, 1994).
Significant QTLs were characterized by their closest associated marker cofactors, LOD score, percentage of explained phenotypic variation, and confidence interval in centimorgans (cM) estimated based on a LOD score drop of 2 on both sides of the likelihood peak.
Genomic positions of the QTLs detected on the linkage groups were drawn using MapChart software (Voorrips RE, 2002) and are illustrated in FIG. 2.
In the F2 population, a QTL explaining 13,3% of the variability between the susceptible and resistant parents is detected on the linkage group 3 for Acidovorax valerianellae resistance. One would note that the linkage group 3 is created and its positions attributed by the mapping software, independently from the Valerianella locusta L. chromosomes or of their physical positions on said chromosomes.
The genomic position of the QTL detected on the Linkage Group 3 (LG 3) of ‘“SRL1 x Susceptible” map by multiple QTL mapping (MQM) for the Acidovorax resistance score noted on leaves is illustrated in FIG. 3 and is summarized below.
The SNPs have been designed with SNP monoplex KASPar technology: KASPar assay used for validation was preformed based on KASP method from KBioscience (LGC Group, Teddington, Middlesex, UK).
Primers for the KASP SNP assays were designed using LGC's primer picker software. For every SNP, two allele-specific forward primers and one common reverse primer per SNP assay were designed. KASP genotyping assays are based on competitive allele-specific PCR and enable bi- allelic scoring of SNPs at specific loci. To summarize, the SNP-specific KASP assay mix and the universal KASP Master mix were added to DNA samples, a thermal cycling reaction was then performed, followed by an end-point fluorescent read. Biallelic discrimination was achieved through the competitive binding of two allele-specific forward primers, each with a unique tail sequence that corresponded with two universal FRET (fluorescence resonant energy transfer) cassettes, one of which was labelled with FAM™ dye and the other of which was labelled with VIC™ dye (LGC, www.lgcgroup. com).
The sequences of the SNPs, including flanking sequences, of the two allele-specific forward primers, of the reverse primer, as well as the allele representative of the susceptible and resistant parent, are given in table 2.
able 2: Sequences of the SNPs. Column 1 reports the name of the SNP, column 2 indicates the position (Pos.) on LG3, column 3 indicates the LOD score, column 4 the sequence of the bi allelic SNP, the sequences of the two allele-specific forward primers are reported in columns 5 and 6 and the sequence of the common reverse primer is in column 7; the allele present in the resistant parent SRL1 and in the deposited seeds is reported in column 8 and the allele of the usceptible parent (Susc. Parent) is reported in the last column.
Example 5: Further marker development and validation
For validation, a set of 74 inbred lines derived from different resistant to moderately resistant F2 from the original mapping population was considered for the validation. The level of resistance of these 74 inbred lines was assessed in a pathologic test following the same protocol than described in example 2. The Acidovorax resistance level is defined as the mean of two separate resistance level measures. Different level of resistance was observed in the set of the 74 inbred lines (see FIG. 4) suggesting the loss of the resistance allele at QTL3 during the fixation process in inbreeds showing low resistance level.
A total of 40 additional SNPs have been genotyped on the panel of 74 inbreds. The SNPs have been selected within the region of QTL3 defined by the interval between markers CS-0019663 and CS- 0015357 (35 cM) which delimit the physical interval containing the QTL.
Each single SNP profile has been compared to the resistance level profile of the 74 tested inbred lines. The marker profiles explaining the better the resistance differences between the 74 inbreds have been retained to constitute a haplotype. The haplotype based on the two markers: CS-0015357 and CS-0022531 explained the most important part of the level of resistance (see FIG. 5).
One would note that CS-0022531 is the best predictive marker for the presence of the QTL conferring the resistance. Its physical position, as well as the physical positions of the most important SNPs on the contig tig00019526 are as follows:
In a second stage of validation, these two most associated SNPs CS-0015357 and CS-0022531 have been validated on a total of 87 lines derived directly or non-directly from the F2 original population and of different generations (F4 to F7 progenies). This material has been phenotyped in patho-test described in example 1 (using a randomized incomplete block design).
Significant effect of QTL3 has been validated through statistical analyses testing the effect of the different QTL3 genotypes (RR, RS and SS) on resistance level, hereafter presented in FIG.6.
“RR” stands for presence (homozygously) of the resistant alleles for both SNPs CS-0015357 and CS-0022531 ,
“RS” stands for presence (heterozygously) of the resistant allele and of the susceptible allele for both SNPs CS-0015357 and CS-0022531 ,
“SS” stands for presence (homozygously) of the susceptible alleles for both SNPs.
Seeds of an F6 progeny, deriving from the F2 original population described in example 2 were collected and deposited at the NCIMB on 17th October 2019, under NCIMB accession number 43500. These seeds are homozygous for the QTL of interest as checked with the SNPs defined above.
Example 6: Genetic Modification of Corn Salad Seeds by Ethyl Methane Sulfonate (EMS)
Seeds of a corn salad varieties are to be treated with EMS by submergence of approximately 2000 seeds per variety into an aerated solution of either 0.5% (w/v) or 0.7% EMS for 24 hours at room temperature.
Approximately 1500 treated seeds per variety per EMS dose are germinated and the resulting plants are grown, preferably in a greenhouse, to produce seeds.
Following maturation, M2 seeds are harvested and bulked in one pool per variety per treatment. The resulting pools of M2 seeds are used as starting material to identify the individual M2 seeds and the plants with an intermediate resistance against A. valerianellae.
REFERENCES:
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics ; 138: 963-971
Elshire et ai, (2011) A robust, simple Genotype-by-sequencing (GBS) Approach for high diversity species. PloS ONE 6(5): e19379.
Gao et ai. (2016). DNA-guided genome editing using the Natronobacterium gregoryi Argonaute. Nature Biotechnology volume 34, pages 768-773.
Gardan L. et ai. (2003) Acidovorax valerianellae sp. nov., a novel pathogen of lamb's lettuce [Valerianella locusta (L.) Laterr] Int J Syst Evol Microbiol. ; 53(Pt 3):795-800 Glaubitz JC, et ai. (2014) TASSEL-GBS: A High Capacity Genotyping by Sequencing Analysis Pipeline. PLoS ONE; 9(2): e90346
Grondeau et ai. (2007) A semiselective medium for the isolation of Acidovorax valerianellae from soil and plant debris. Plant Pathology; 56(2):302-310.
Grondeau C ; Samson R. (2009) Detection of Acidovorax valerianellae in corn-salad seeds, seed transmission of the pathogen and disease development in the field . Plant Pathology; 58, 846-852 Jansen RC and Stam P (1994) High Resolution of Quantitative Traits into Multiple Loci via Interval Mapping. Genetics 136: 1447-1455
Kenis K, Keulemans J (2007) Study of tree architecture of apple (Malus 6 domestica Borkh) by QTL analysis of growth traits. Molecular Breeding 19: 193-208.
Kosambi DD (1944) The estimation of map distance from recombination values. Annals of Eugenics 12: 172-175.
Muminovic J. et ai. (2004) Genetic diversity in cornsalad (Valerianella locusta) and related species as determined by AFLP markers. Plant Breeding 123: 460-466.
Rhoads A, Au KF (2015). PacBio Sequencing and Its Applications. Genomics proteomics Bioinformatics 13: 278-289.
Stam P. (1993). Construction of integrated genetic linkage maps by means of a new computer package : JoinMap. The Plant Journal 3: 739-744.
Van Ooijen JW (2009) MapQTL 6, Software for the mapping of quantitative trait loci in experimental populations of diploid species. Kyasma BV, Wageningen, Netherlands Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93: 77-8;
Vos P. et al. (1995). AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res.
23(21 ):4407-14
Claims
1. Corn salad plant resistant to Acidovorax valerianellae comprising in its genome a quantitative trait locus (QTL), wherein said QTL confers to the plant resistance to Acidovorax valerianellae and wherein said QTL can be identified with the molecular marker CS-0022531 (SEQ ID NO:14).
2. Plant according to claim 1 , wherein said QTL can be identified by allele A of CS-0022531.
3. Plant according to claim 1 or 2, wherein said QTL can be identified by the haplotype corresponding to allele A of CS-0022531 (SEQ ID NO: 14) and allele A of CS-0015357 (SEQ ID NO:9).
4. A plant according to any one of claims 1 to 3, wherein said QTL is present in the genome of the seeds of plant ValeriacidoR, NCIMB accession number 43500.
5. The plant according to any one of claims 1 to 4, wherein said plant is a Valerianella locusta L. plant, a Valerianella locusta L. locusta plant or Valerianella locusta.
6. The plant according to any one of claims 1 to 5, wherein said plant is a progeny of the plant ValeriacidoR, seeds thereof have been deposited at the NCIMB, under NCIMB accession number 43500.
7. The plant according to any one of claims 1 to 6, wherein said plant is an edible, commercial, cultivated corn salad plant.
8. The plant according to any one of claims 1 to 7, wherein said QTL is located within the genomic interval delimited by CS-0019663 (SEQ ID NO:7) and CS-0021333 (SEQ ID NO:10), preferably within a genomic interval delimited by CS-16322 (SEQ ID NO:8) and CS-0015357 (SEQ ID NO:9), or overlaps said interval.
9. The plant according to any one of claims 1 to 8, wherein said QTL can be detected by one or more of the following molecular markers: CS-0022531 (SEQ ID NO:14), CS-0015357 (SEQ ID NO:9) and/or CS-0016322 (SEQ ID NO:8).
10. A cell of a V. locusta plant according to any one of claims 1-9, comprising in its genome said QTL, conferring resistance against A. valerianellae.
11. A cell according to claim 10, wherein said cell is a regenerable cell, or a non regenerable cell.
12. Plant part of a plant according to any one of claims 1-9, in particular seeds, explants, reproductive material, propagation material, scion, cutting, seed, fruit, root, rootstock, microspore, pollen, ovule, embryo, protoplast, leaf, anther, stem, petiole or flowers, wherein said plant part comprises cells according to claim 10.
13. Seed of a plant, which develops into a plant according to any one of claims 1-9.
14. A tissue culture of cells of the plant according to any one of claims 1-9, wherein the cells are derived from embryos, protoplasts, meristematic cells, callus, pollen, leaves, anthers, stems, petioles, roots, root tips, seeds, flowers, cotyledons, and/or hypocotyls, and contain in their genome said QTL conferring resistance against A. valerianellae, preferably homozygously.
15. Use of a plant or seed of V. locusta, deposited at the NCIMB under the accession number NCIMB 43500, or progeny thereof, bearing homozygously the QTL as defined in claim 1 conferring resistance against A. valerianellae infection, as a breeding partner in a breeding program for conferring said resistance to V. locusta plants, preferably to V. locusta plants susceptible to A. valerianellae.
16. A method for breeding V. locusta plants having resistance against A. valerianellae , comprising the steps of crossing a plant grown from the deposited seeds NCIMB 43500 or progeny thereof bearing a QTL conferring A. valerianellae resistance, with an initial V. locusta plant devoid of said QTL, wherein said QTL is present in the genome of the seeds of plant ValeriacidoR, NCIMB accession number 43500, and is identifiable by allele A of CS-0022531 or by the haplotype corresponding to allele A of CS-0022531 and allele A of CS-0015357.
17. A method for conferring resistance against A. valerianellae to a V. locusta plant, comprising genetically modifying said plant to introduce a QTL conferring said resistance, wherein said QTL is identifiable by allele A of CS-0022531 , or by the haplotype corresponding to allele A of CS-0022531 and allele A of CS-0015357, and is present in the genome of the seeds of plant ValeriacidoR, NCIMB accession number 43500.
18. A method for conferring resistance against A. valerianellae to V. locusta plants, comprising the steps of: a) Crossing a plant grown from the deposited seeds NCIMB 43500, or progeny thereof, bearing the QTL conferring A. valerianellae resistance, and an initial V. locusta plant preferably devoid of said QTL; b) Selecting a plant in the progeny thus obtained, bearing the QTL; c) Optionally self-pollinating one or several times the plant obtained at step b) and selecting in the progeny thus obtained a plant having A. valerianellae resistance,
wherein said QTL is present in the genome of the seeds of plant ValeriacidoR, NCIMB accession number 43500.
19. A method for conferring resistance against A. valerianellae to V. locusta plants, comprising the steps of: a1) Crossing a plant grown from the deposited seeds NCIMB 43500, or progeny thereof, bearing the QTL conferring A. valerianellae resistance and an initial V. locusta plant, preferably devoid of said QTL, thus generating F1 hybrids, a2) Selfing the F1 hybrids to create F2 population, b) Selecting individuals in the progeny thus obtained having resistance against A. valerianellae , wherein said QTL is present in the genome of the seeds of plant ValeriacidoR, NCIMB accession number 43500.
20. The method of claim 18 or 19, wherein SNPs markers are used in steps b) and/or c) for selecting plants bearing the QTL conferring A. valerianellae resistance.
21. A method for detecting and/or selecting corn salad plant comprising a QTL conferring A. valerianellae resistance, wherein said method comprises the detection of at least one genetic marker linked to the QTL, wherein said genetic marker is selected in the group comprising CS- 0022531 , CS-0015357 and/or CS-0016322.
22. A method for detecting and/or selecting a corn salad plant having a QTL, conferring A. valerianellae resistance, said method comprising the detection of the following alleles: allele A of CS-0022531 , allele A of CS-0015357 or allele A of CS-0016322, in a genetic material sample of the plant to be selected, and preferably the haplotype corresponding to allele A of CS-0022531 and allele A of CS-0015357.
23. A method for detecting and/or selecting corn salad plants having a QTL, conferring A. valerianellae resistance, said method comprising: a) Assaying corn salad plants for the presence of at least one genetic marker, genetically linked to the QTL conferring A. valerianellae resistance in corn salad, b) Selecting a plant comprising the genetic marker and the QTL conferring A. valerianellae resistance, wherein the QTL and the genetic marker are to be found in the genomic interval delimited by CS-0019663 and CS-0021333.
24. Use of a genetic marker for detecting the presence of sequences conferring A. valerianellae resistance, wherein said genetic marker is selected from the group comprising CS-0022531 , CS-0015357 and/or CS-0016322.
25. Method for identifying a molecular marker associated with the QTL conferring A. valerianellae resistance in corn salad, comprising: identifying a molecular marker in the genomic interval delimited by CS-0019663 and CS- 0021333 or at less than 2 megabase units from the locus of CS-0016322, CS-0022531 or CS-0015357, preferably less than 0.5 megabase units; and determining whether an allele or state of said molecular marker is associated with the phenotype of A. valerianellae resistance in a segregating population issued from a plant exhibiting A. valerianellae resistance.
26. Use of at least one of the SNP markers CS-0016322, CS-0022531 and CS-0015357 associated with a QTL conferring A. valerianellae resistance to corn salad, for identifying an alternative molecular marker associated with said QTL, wherein said alternative molecular marker is in the genomic interval delimited by CS-0019663 and CS-0021333, or at less than 2 megabase units from the locus of CS-0016322, CS-0022531 or CS-0015357, preferably less than 0.5 megabase units.
27. A method for improving the yield of corn salad plant in an environment infected by A. valerianellae comprising growing corn salad comprising in its genome a QTL conferring to said plant A. valerianellae resistance, wherein said QTL is present in the genome of a plant of the seeds ValeriacidoR NCIMB accession number 43500 and is identifiable by allele A of CS- 0022531 or by the haplotype corresponding to allele A of CS-0022531 and allele A of CS- 0015357.
28. A method of protecting a field, tunnel, greenhouse or glasshouse of corn salad from A. valerianellae infection, comprising growing resistant corn salad comprising in its genome a QTL, conferring to said corn salad resistance against A. valerianellae , wherein said QTL is present in the genome of a plant of the seeds ValeriacidoR NCIMB accession number 43500 and is identifiable by allele A of CS-0022531 or by the haplotype corresponding to allele A of CS-0022531 and allele A of CS-0015357.
29. The method according to any one of claims 27-28, wherein said corn salad plants are plants according to any one of claims 1-9.
30. Use of a corn salad plant having resistance to A. valerianellae for limiting or controlling A. valerianellae infection of a field, tunnel, greenhouse or glasshouse, wherein said corn salad plant comprises in its genome a QTL conferring to said plant A. valerianellae resistance, wherein said QTL is present in the genome of a plant of the seeds ValeriacidoR NCIMB accession number 43500 and is identifiable by the haplotype corresponding to allele A of CS- 0022531 (SEQ ID NO:14) and allele A of CS-0015357 (SEQ ID NO:9).
Priority Applications (2)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| EP19838940.5A EP4057802A1 (en) | 2019-11-14 | 2019-11-14 | Resistance to acidovorax valerianellae in corn salad |
| PCT/IB2019/001290 WO2021094805A1 (en) | 2019-11-14 | 2019-11-14 | Resistance to acidovorax valerianellae in corn salad |
Applications Claiming Priority (1)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| PCT/IB2019/001290 WO2021094805A1 (en) | 2019-11-14 | 2019-11-14 | Resistance to acidovorax valerianellae in corn salad |
Publications (1)
| Publication Number | Publication Date |
|---|---|
| WO2021094805A1 true WO2021094805A1 (en) | 2021-05-20 |
Family
ID=69173090
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| PCT/IB2019/001290 WO2021094805A1 (en) | 2019-11-14 | 2019-11-14 | Resistance to acidovorax valerianellae in corn salad |
Country Status (2)
| Country | Link |
|---|---|
| EP (1) | EP4057802A1 (en) |
| WO (1) | WO2021094805A1 (en) |
Citations (10)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US8586363B2 (en) | 2009-12-10 | 2013-11-19 | Regents Of The University Of Minnesota | TAL effector-mediated DNA modification |
| WO2014009555A1 (en) | 2012-07-13 | 2014-01-16 | Rijk Zwaan Zaadteelt En Zaadhandel B.V. | Small seeded corn salad (valerianella locusta) |
| US8697359B1 (en) | 2012-12-12 | 2014-04-15 | The Broad Institute, Inc. | CRISPR-Cas systems and methods for altering expression of gene products |
| US8795965B2 (en) | 2012-12-12 | 2014-08-05 | The Broad Institute, Inc. | CRISPR-Cas component systems, methods and compositions for sequence manipulation |
| US8865406B2 (en) | 2012-12-12 | 2014-10-21 | The Broad Institute Inc. | Engineering and optimization of improved systems, methods and enzyme compositions for sequence manipulation |
| US8889356B2 (en) | 2012-12-12 | 2014-11-18 | The Broad Institute Inc. | CRISPR-Cas nickase systems, methods and compositions for sequence manipulation in eukaryotes |
| US8906616B2 (en) | 2012-12-12 | 2014-12-09 | The Broad Institute Inc. | Engineering of systems, methods and optimized guide compositions for sequence manipulation |
| US8993233B2 (en) | 2012-12-12 | 2015-03-31 | The Broad Institute Inc. | Engineering and optimization of systems, methods and compositions for sequence manipulation with functional domains |
| US9145565B2 (en) | 2002-01-23 | 2015-09-29 | University Of Utah Research Foundation | Targeted chromosomal mutagenesis using zinc finger nucleases |
| US9181535B2 (en) | 2012-09-24 | 2015-11-10 | The Chinese University Of Hong Kong | Transcription activator-like effector nucleases (TALENs) |
-
2019
- 2019-11-14 WO PCT/IB2019/001290 patent/WO2021094805A1/en unknown
- 2019-11-14 EP EP19838940.5A patent/EP4057802A1/en active Pending
Patent Citations (16)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US9145565B2 (en) | 2002-01-23 | 2015-09-29 | University Of Utah Research Foundation | Targeted chromosomal mutagenesis using zinc finger nucleases |
| US8586363B2 (en) | 2009-12-10 | 2013-11-19 | Regents Of The University Of Minnesota | TAL effector-mediated DNA modification |
| WO2014009555A1 (en) | 2012-07-13 | 2014-01-16 | Rijk Zwaan Zaadteelt En Zaadhandel B.V. | Small seeded corn salad (valerianella locusta) |
| US9181535B2 (en) | 2012-09-24 | 2015-11-10 | The Chinese University Of Hong Kong | Transcription activator-like effector nucleases (TALENs) |
| US8895308B1 (en) | 2012-12-12 | 2014-11-25 | The Broad Institute Inc. | Engineering and optimization of improved systems, methods and enzyme compositions for sequence manipulation |
| US8865406B2 (en) | 2012-12-12 | 2014-10-21 | The Broad Institute Inc. | Engineering and optimization of improved systems, methods and enzyme compositions for sequence manipulation |
| US8871445B2 (en) | 2012-12-12 | 2014-10-28 | The Broad Institute Inc. | CRISPR-Cas component systems, methods and compositions for sequence manipulation |
| US8889356B2 (en) | 2012-12-12 | 2014-11-18 | The Broad Institute Inc. | CRISPR-Cas nickase systems, methods and compositions for sequence manipulation in eukaryotes |
| US8795965B2 (en) | 2012-12-12 | 2014-08-05 | The Broad Institute, Inc. | CRISPR-Cas component systems, methods and compositions for sequence manipulation |
| US8906616B2 (en) | 2012-12-12 | 2014-12-09 | The Broad Institute Inc. | Engineering of systems, methods and optimized guide compositions for sequence manipulation |
| US8932814B2 (en) | 2012-12-12 | 2015-01-13 | The Broad Institute Inc. | CRISPR-Cas nickase systems, methods and compositions for sequence manipulation in eukaryotes |
| US8945839B2 (en) | 2012-12-12 | 2015-02-03 | The Broad Institute Inc. | CRISPR-Cas systems and methods for altering expression of gene products |
| US8993233B2 (en) | 2012-12-12 | 2015-03-31 | The Broad Institute Inc. | Engineering and optimization of systems, methods and compositions for sequence manipulation with functional domains |
| US8999641B2 (en) | 2012-12-12 | 2015-04-07 | The Broad Institute Inc. | Engineering and optimization of systems, methods and compositions for sequence manipulation with functional domains |
| US8771945B1 (en) | 2012-12-12 | 2014-07-08 | The Broad Institute, Inc. | CRISPR-Cas systems and methods for altering expression of gene products |
| US8697359B1 (en) | 2012-12-12 | 2014-04-15 | The Broad Institute, Inc. | CRISPR-Cas systems and methods for altering expression of gene products |
Non-Patent Citations (19)
| Title |
|---|
| CHURCHILL GADOERGE RW: "Empirical threshold values for quantitative trait mapping", GENETICS, vol. 138, 1994, pages 963 - 971 |
| ELSHIRE ET AL.: "A robust, simple Genotype-by-sequencing (GBS) Approach for high diversity species", PLOS ONE, vol. 6, no. 5, 2011, pages e19379, XP055033060, DOI: 10.1371/journal.pone.0019379 |
| ESTELLE NODODUS: "Dépérissement de la mâche (Valerianella locusta) en région nantaise : étiologie du dépérissement, caractérisation des Pythiacées impliquées et recherche de premières options de protection", 4 November 2016 (2016-11-04), XP055678012, Retrieved from the Internet <URL:https://dumas.ccsd.cnrs.fr/dumas-01391964/document> [retrieved on 20200319] * |
| GAO ET AL.: "DNA-guided genome editing using the Natronobacterium gregoryi Argonaute", NATURE BIOTECHNOLOGY, vol. 34, 2016, pages 768 - 773, XP055518128, DOI: 10.1038/nbt.3547 |
| GARDAN L. ET AL.: "Acidovorax valerianellae sp. nov., a novel pathogen of lamb's lettuce [Valerianella locusta (L.) Laterr]", INT J SYST EVOL MICROBIOL., vol. 53, 2003, pages 795 - 800 |
| GLAUBITZ JC ET AL.: "TASSEL-GBS: A High Capacity Genotyping by Sequencing Analysis Pipeline", PLOS ONE, vol. 9, no. 2, 2014, pages e90346 |
| GRONDEAU CSAMSON R: "Detection of Acidovorax valerianellae in corn-salad seeds, seed transmission of the pathogen and disease development in the field", PLANT PATHOLOGY, vol. 58, 2009, pages 846 - 852 |
| GRONDEAU ET AL.: "A semiselective medium for the isolation of Acidovorax valerianellae from soil and plant debris", PLANT PATHOLOGY, vol. 56, no. 2, 2007, pages 302 - 310, XP055037934, DOI: 10.1111/j.1365-3059.2006.01510.x |
| JANSEN RCSTAM P: "High Resolution of Quantitative Traits into Multiple Loci via Interval Mapping", GENETICS, vol. 136, 1994, pages 1447 - 1455, XP055250519 |
| KENIS KKEULEMANS J: "Study of tree architecture of apple (Malus 6 domestica Borkh) by QTL analysis of growth traits", MOLECULAR BREEDING, vol. 19, 2007, pages 193 - 208, XP019482973, DOI: 10.1007/s11032-006-9022-5 |
| KOSAMBI DD: "The estimation of map distance from recombination values", ANNALS OF EUGENICS, vol. 12, 1944, pages 172 - 175 |
| L. GARDAN: "Acidovorax valerianellae sp. nov., a novel pathogen of lamb's lettuce [Valerianella locusta (L.) Laterr.]", INTERNATIONAL JOURNAL OF SYSTEMATIC AND EVOLUTIONARY MICROBIOLOGY, vol. 53, no. 3, 1 May 2003 (2003-05-01), GB, pages 795 - 800, XP055678005, ISSN: 1466-5026, DOI: 10.1099/ijs.0.02404-0 * |
| MUMINOVIC J ET AL.: "Genetic diversity in cornsalad (Valerianella locusta) and related species as determined by AFLP markers", PLANT BREEDING, vol. 123, 2004, pages 460 - 466, XP002580359, DOI: 10.1111/j.1439-0523.2004.tb01854.x |
| MUMINOVIC J ET AL: "Genetic diversity in cornsalad (Valerianella locusta) and related species as determined by AFLP markers", PLANT BREEDING, PAUL PAREY SCIENTIFIC PUBL., BERLIN, DE, vol. 123, no. 5, 1 October 2004 (2004-10-01), pages 460 - 466, XP002580359, ISSN: 0179-9541, [retrieved on 20080128], DOI: 10.1111/J.1439-0523.2004.TB01854.X * |
| RHOADS AAU KF: "PacBio Sequencing and Its Applications", GENOMICS PROTEOMICS BIOINFORMATICS, vol. 13, 2015, pages 278 - 289, XP029335898, DOI: 10.1016/j.gpb.2015.08.002 |
| STAM P.: "Construction of integrated genetic linkage maps by means of a new computer package: JoinMap", THE PLANT JOURNAL, vol. 3, 1993, pages 739 - 744, XP003005041 |
| VAN OOIJEN JW: "MapQTL 6, Software for the mapping of quantitative trait loci in experimental populations of diploid species", 2009, KYASMA BV |
| VOORRIPS RE: "MapChart: software for the graphical presentation of linkage maps and QTLs", J HERED, vol. 93, 2002, pages 77 - 8 |
| VOS P. ET AL.: "AFLP: a new technique for DNA fingerprinting", NUCLEIC ACIDS RES., vol. 23, no. 21, 1995, pages 4407 - 14 |
Also Published As
| Publication number | Publication date |
|---|---|
| EP4057802A1 (en) | 2022-09-21 |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| US11889812B2 (en) | Tolerance in plants of Solanum lycopersicum to the tobamovirus tomato brown rugose fruit virus (TBRFV) | |
| US10939628B2 (en) | Resistance to ToLCNDV in melons | |
| US11730135B2 (en) | Resistance in plants of Solanum lycopersicum to the tobamovirus tomato brown rugose fruit virus | |
| AU2016305521B2 (en) | Resistance to Australian variant of A. Candida race 9 in broccoli | |
| US20210307275A1 (en) | Resistance to xanthomonas campestris pv. campestris (xcc) in cauliflower | |
| US20220279748A1 (en) | Tolerance to tolcndv in cucumber | |
| US20230276763A1 (en) | Resistance in plants of solanum lycopersicum to the tobrfv | |
| US20220304272A1 (en) | Resistance in plants of solanum lycopersicum to the tobamovirus tomato brown rugose fruit virus | |
| US11277983B2 (en) | Peronospora resistance in spinacia sp | |
| WO2021094805A1 (en) | Resistance to acidovorax valerianellae in corn salad | |
| EP4256950A1 (en) | Tolerance to cgmmv in cucumber | |
| US11365427B2 (en) | Spinacia oleraceae plant resistant to Albugo occidentalis and Peronospora farinosa | |
| WO2024223900A1 (en) | Genetic determinants conferring improved tobrfv resistance | |
| WO2024223901A1 (en) | Resistance to tomato brown rugose fruit virus at elevated temperatures | |
| EP4380353A1 (en) | Resistance to leveillula taurica in pepper | |
| WO2017025627A1 (en) | Resistance to australian variant of a. candida race 9 in broccoli | |
| KR20240153272A (en) | Spinach line msa-s021-1336m |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| 121 | Ep: the epo has been informed by wipo that ep was designated in this application |
Ref document number: 19838940 Country of ref document: EP Kind code of ref document: A1 |
|
| NENP | Non-entry into the national phase |
Ref country code: DE |
|
| ENP | Entry into the national phase |
Ref document number: 2019838940 Country of ref document: EP Effective date: 20220614 |