WO2017148432A1 - 用于预测慢性乙肝患者对IFNα治疗的应答的方法和试剂盒 - Google Patents
用于预测慢性乙肝患者对IFNα治疗的应答的方法和试剂盒 Download PDFInfo
- Publication number
- WO2017148432A1 WO2017148432A1 PCT/CN2017/075542 CN2017075542W WO2017148432A1 WO 2017148432 A1 WO2017148432 A1 WO 2017148432A1 CN 2017075542 W CN2017075542 W CN 2017075542W WO 2017148432 A1 WO2017148432 A1 WO 2017148432A1
- Authority
- WO
- WIPO (PCT)
- Prior art keywords
- sample
- marker
- subject
- ifnα
- pbmc
- Prior art date
Links
Images
Classifications
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N15/00—Mutation or genetic engineering; DNA or RNA concerning genetic engineering, vectors, e.g. plasmids, or their isolation, preparation or purification; Use of hosts therefor
- C12N15/09—Recombinant DNA-technology
- C12N15/11—DNA or RNA fragments; Modified forms thereof; Non-coding nucleic acids having a biological activity
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12Q—MEASURING OR TESTING PROCESSES INVOLVING ENZYMES, NUCLEIC ACIDS OR MICROORGANISMS; COMPOSITIONS OR TEST PAPERS THEREFOR; PROCESSES OF PREPARING SUCH COMPOSITIONS; CONDITION-RESPONSIVE CONTROL IN MICROBIOLOGICAL OR ENZYMOLOGICAL PROCESSES
- C12Q1/00—Measuring or testing processes involving enzymes, nucleic acids or microorganisms; Compositions therefor; Processes of preparing such compositions
- C12Q1/70—Measuring or testing processes involving enzymes, nucleic acids or microorganisms; Compositions therefor; Processes of preparing such compositions involving virus or bacteriophage
- C12Q1/701—Specific hybridization probes
- C12Q1/706—Specific hybridization probes for hepatitis
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12Q—MEASURING OR TESTING PROCESSES INVOLVING ENZYMES, NUCLEIC ACIDS OR MICROORGANISMS; COMPOSITIONS OR TEST PAPERS THEREFOR; PROCESSES OF PREPARING SUCH COMPOSITIONS; CONDITION-RESPONSIVE CONTROL IN MICROBIOLOGICAL OR ENZYMOLOGICAL PROCESSES
- C12Q1/00—Measuring or testing processes involving enzymes, nucleic acids or microorganisms; Compositions therefor; Processes of preparing such compositions
- C12Q1/68—Measuring or testing processes involving enzymes, nucleic acids or microorganisms; Compositions therefor; Processes of preparing such compositions involving nucleic acids
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12Q—MEASURING OR TESTING PROCESSES INVOLVING ENZYMES, NUCLEIC ACIDS OR MICROORGANISMS; COMPOSITIONS OR TEST PAPERS THEREFOR; PROCESSES OF PREPARING SUCH COMPOSITIONS; CONDITION-RESPONSIVE CONTROL IN MICROBIOLOGICAL OR ENZYMOLOGICAL PROCESSES
- C12Q2600/00—Oligonucleotides characterized by their use
- C12Q2600/106—Pharmacogenomics, i.e. genetic variability in individual responses to drugs and drug metabolism
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12Q—MEASURING OR TESTING PROCESSES INVOLVING ENZYMES, NUCLEIC ACIDS OR MICROORGANISMS; COMPOSITIONS OR TEST PAPERS THEREFOR; PROCESSES OF PREPARING SUCH COMPOSITIONS; CONDITION-RESPONSIVE CONTROL IN MICROBIOLOGICAL OR ENZYMOLOGICAL PROCESSES
- C12Q2600/00—Oligonucleotides characterized by their use
- C12Q2600/158—Expression markers
Definitions
- the present invention relates to markers for predicting the response of chronic hepatitis B patients to IFN[alpha] treatment.
- the invention also relates to a method for predicting the response of a chronic hepatitis B patient to IFN[alpha] treatment comprising the step of determining the level of expression of such a marker in PBMC of a chronic hepatitis B patient.
- the invention also relates to a kit for use in the above method.
- Hepatitis B virus infection is one of the most important public health problems in the world. There are currently more than 350 million chronic hepatitis B virus infections worldwide. Chronic hepatitis B virus infection can cause chronic liver disease such as Chronic hepatitis B (CHB), Liver cirrhosis (LC) and Hepatocellular carcinoma (HCC). Hepatitis virus infection and related diseases caused by death are more than 1 million people worldwide each year.
- CHB Chronic hepatitis B
- LC Liver cirrhosis
- HCC Hepatocellular carcinoma
- Chronic HBV infection is a dynamic process, and the natural history of infection can be divided into four different stages, which are not necessarily inextricably linked: 1) immune tolerance stage, characterized by HBeAg positive, active HBV DNA replication, ALT The level is normal or low, generally no fibrosis; 2) active immune stage / immune clearance stage, characterized by HBeAg positive but its level begins to decline, HBV DNA replication level decreases, serum HBsAg levels decrease, ALT continues to rise or fluctuation.
- HBeAg-negative hepatitis which belongs to the late stage of chronic hepatitis B, is characterized by periodic fluctuations in HBV DNA and ALT levels and active hepatitis.
- the HBV virus which was previously variant of the C region and/or the basic core promoter (BCP), is predominantly unable to express or express very low levels of HBeAg.
- HBV virus has no direct pathogenic effect on host cells, and its main pathogenic mechanism is that HBV virus causes host immune response, and immune response causes immune liver tissue damage.
- the treatment of chronic HBV infection requires different antiviral treatments depending on the individual patient.
- the purpose of antiviral therapy is to continue to suppress HBV virus, Prevent the development or transformation of hepatitis into fibrosis, cirrhosis and even liver cancer, prolong the survival of patients and improve the quality of life.
- nucleoside analogues lamivudine (LAM), telbivudine (LdT), emtricitabine (FTC), entecavir (ETV), Defovir (ADV), tenofovir (TDF); two interferons: common interferon (IFN ⁇ ) and pegylated interferon (PEG-IFN ⁇ ).
- LAM lamivudine
- LdT telbivudine
- FTC emtricitabine
- ETV entecavir
- ADV Defovir
- TDF tenofovir
- IFN ⁇ common interferon
- PEG-IFN ⁇ pegylated interferon
- IFN ⁇ was first approved by the FDA for the treatment of chronic hepatitis B.
- PEG-IFN ⁇ is also the drug of choice for the treatment of chronic hepatitis B, with antiviral and immunomodulatory effects. effect.
- the main advantage of IFN ⁇ is its non-resistance and immune-mediated control of HBV infection, resulting in a higher chance of HBeAg seroconversion, a more sustained virological response, and HBsAg clearance at the end of treatment.
- the level of HBV DNA in the medium is maintained at the lower limit of detection.
- Antiviral therapy must ensure a certain degree of virological suppression and can produce biochemical remission, histological recovery and prevention of complications.
- the ideal end point for HBV treatment is the disappearance of HBsAg, but existing antiviral drugs are difficult to achieve this goal, so a more realistic treatment endpoint is the ability to induce a sustained virological suppression state.
- Antiviral responses can be divided into biochemical levels, serum levels, virological levels, and histological levels. All responses can be assessed during and after treatment. Clinical studies have shown that the response rate of CHB patients treated with IFN ⁇ for 6 months is only 25-40%. Determining the ability of CHB patients to respond to HBV-specific immune responses may be able to predict the expected efficacy of CHB patients receiving treatment.
- markers that are capable of predicting the response of CHB patients to IFN[alpha] treatment in a simple, convenient, rapid, and highly accurate and specific manner.
- IFN ⁇ or " ⁇ -type interferon” as used herein includes all natural or recombinant alpha-type interferons, particularly preferably human alpha-type interferons, such as recombinant human alpha-type interferons, including but not limited to IFN ⁇ -1b (eg available from Schering Corporation, Kenilworth, NJ) Interferon), IFN ⁇ -2a (eg available from Hoffmann-La Roche, Nutley, NJ) Interferon) or IFN ⁇ -2b (eg available from Schering Corporation, Kenilworth, NJ) Interferon); for example a mixture of natural alpha interferons, including but not limited to IFN ⁇ -n1 (eg available from Sumitomo, Japan) Or Glaxo-Wellcome Ltd., London, Great Britain Interferon alpha-n1) or IFNa-n3 (eg available from Alferon of Interferon Sciences) Interferon).
- IFN ⁇ -1b eg available from Schering Corporation, Kenilworth, NJ
- the term “IFN ⁇ ” or “ ⁇ -type interferon” also includes any substance having biological activity of IFN ⁇ , such as a mutant or modified IFN ⁇ , such as a PEG derivative of IFN ⁇ (PEG-IFN ⁇ ).
- the term “IFN ⁇ ” or “ ⁇ -type interferon” is not limited by any particular source of acquisition, may be obtained from commercially available sources or may be produced by conventional techniques known to those skilled in the art, including However, it is not limited to the biological source extraction method and the genetic engineering extraction method, which are described in detail, for example, in “Pestka S. Arch Biochem Biophys. 1983 Feb 15; 221(1): 1-37” (which is incorporated herein by reference).
- HBV antigen refers to a protein present in hepatitis B virus (HBV) that is capable of eliciting an immune response in the body, including hepatitis B virus core antigen (HBcAg), hepatitis B virus surface. Antigen (HBsAg) and hepatitis B virus E antigen (HBeAg).
- HBV hepatitis B virus
- HBcAg when referring to the amino acid sequence of HBcAg, the description is made with reference to the sequence shown in SEQ ID NO:38.
- mutations or mutations including but not limited to, substitutions, deletions and/or additions, such as different genotypes, subtypes or different serotypes
- HBcAg shall include all such sequences, including, for example, the sequences set forth in SEQ ID NO: 38, as well as natural or artificial variants thereof.
- sequence fragment of HBcAg when describing a sequence fragment of HBcAg, it includes not only the sequence fragment of SEQ ID NO: 38 but also the corresponding sequence fragment in its native or artificial variant.
- amino acid residues 1-183 of HBcAg includes the amino acid residues 1-183 of SEQ ID NO: 38, and the corresponding fragments thereof (natural or artificial).
- corresponding segment means a segment located at an equivalent position in the sequence to be compared when the sequences are optimally aligned, that is, when the sequences are aligned to obtain the highest percentage identity.
- HBecAg is not limited by any particular method of synthesizing a protein, and can be produced by conventional techniques known to those skilled in the art, such as DNA recombination techniques or chemical synthesis techniques.
- the expression "antigenic fragment of HBcAg” refers to a fragment of an amino acid sequence (ie, a polypeptide) obtained by truncating an HBcAg protein, the fragment having the same biological activity as the corresponding full-length protein, That is, PBMC from chronic hepatitis B patients can be stimulated and activated.
- the amino acid sequence fragment shown by SEQ ID NO: 3-37 in the present invention is an antigenic fragment of HBcAg.
- the antigenic fragment is not limited by any particular method of synthesizing the polypeptide, and can be produced by conventional techniques known to those skilled in the art, such as DNA recombination techniques or chemical synthesis techniques.
- HBcAg or an antigenic fragment thereof can be obtained by a DNA recombination technique, for example, by encoding using a cell-free expression system.
- Cell-free expression systems include, for example, reticulocyte-based lysate-based expression systems, wheat germ extract-based expression systems, and E. coli extract-based expression systems); or by using in vivo expression Systems (eg, E. coli prokaryotic expression systems, yeast eukaryotic expression systems) are obtained from polynucleotides encoding these proteins or polypeptides.
- HBcAg or an antigenic fragment thereof can be produced by chemical synthesis.
- Methods for chemical total synthesis of proteins or polypeptides are well known in the art (see, for example, Raibaut L, et al., Top Curr Chem. 2015; 363: 103-54; Thapa P, et al. Molecules.
- SPPS Solid Phase Peptide Synthesis
- NCL Native Chemical Ligation
- TAEC Transfer Active Ester Condensation
- an antigenic fragment having an amino acid sequence as shown in SEQ ID NO: 3-37, respectively means an antigenic fragment having the amino acid sequence of SEQ ID NO: 3, having SEQ An antigenic fragment of the amino acid sequence shown by ID NO: 4, a combination of an antigenic fragment having the amino acid sequence of SEQ ID NO: 36, and an antigenic fragment having the amino acid sequence of SEQ ID NO: 37.
- immunological assay refers to an assay that utilizes specific interaction/binding affinity between antigen-antibodies, which is generally useful for detecting the presence of a particular antigen or antibody in a sample or Level.
- immunological assays are well known to those skilled in the art and include, but are not limited to, ELISA assays, Elispot assays Measurement, Western blotting, surface plasmon resonance, and the like.
- ELISA assays Elispot assays Measurement, Western blotting, surface plasmon resonance, and the like.
- antibody refers to an immunoglobulin molecule that is typically composed of two pairs of polypeptide chains, each pair having a "light” (L) chain and a “heavy” (H) chain.
- Antibody light chains can be classified as K and lambda light chains.
- Heavy chains can be classified as ⁇ , ⁇ , ⁇ , ⁇ , or ⁇ , and the isotypes of antibodies are defined as IgM, IgD, IgG, IgA, and IgE, respectively.
- the variable and constant regions are joined by a "J" region of about 12 or more amino acids, and the heavy chain further comprises a "D" region of about 3 or more amino acids.
- Each heavy chain consists of a heavy chain variable region (VH) and a heavy chain constant region (CH).
- the heavy chain constant region consists of three domains (CH1, CH2 and CH3).
- Each light chain consists of a light chain variable region (VL) and a light chain constant region (CL).
- the light chain constant region consists of one domain CL.
- the constant region of the antibody mediates binding of the immunoglobulin to host tissues or factors, including various cells of the immune system (eg, effector cells) and the first component (Clq) of the classical complement system.
- the VH and VL regions can also be subdivided into regions with high denaturation (referred to as complementarity determining regions (CDRs)) interspersed with more conserved regions called framework regions (FR).
- CDRs complementarity determining regions
- Each VH and VL consists of three CDRs and four FRs arranged in the following order: FR1, CDR1, FR2, CDR2, FR3, CDR3, FR4 from the amino terminus to the carboxy terminus.
- the variable regions (VH and VL) of each heavy/light chain pair form an antibody binding site, respectively.
- the assignment of amino acids to regions or domains follows the Kabat Sequences of Proteins of Immunological Interest (National Institutes of Health, Bethesda, Md. (1987 and 1991)), or Chothia & Lesk (1987) J. Mol. Biol. 196:901-917; Chothia et al. (1989) Nature 342: 878-883.
- the term "antibody” is not limited by any particular method of producing antibodies.
- the antibodies may be antibodies of different isotypes, for example, IgG (eg, IgGl, IgG2, IgG3 or IgG4 subtype), IgA1, IgA2, IgD, IgE or IgM antibodies.
- IgG eg, IgGl, IgG2, IgG3 or IgG4 subtype
- IgA1, IgA2, IgD, IgE or IgM antibodies for example, IgG (eg, IgGl, IgG2, IgG3 or IgG4 subtype), IgA1, IgA2, IgD, IgE or IgM antibodies.
- an "antigen-binding fragment" of an antibody refers to one or more portions of a full length antibody that retain the ability to bind to the same antigen (eg, OAS2 or USP18) to which the antibody binds, capable of Intact antibodies compete for specific binding to antigen. See generally, Fundamental Immunology, Ch. 7 Paul, W., ed., 2nd Ed., Raven Press, N. Y. (1989), which is incorporated herein by reference in its entirety for all purposes.
- Antigen-binding fragments can be produced by recombinant DNA techniques or by enzymatic or chemical cleavage of intact antibodies.
- antigen-binding fragments include Fab, Fab', F(ab')2, Fd, Fv, dAb and complementarity determining region (CDR) fragments, single chain antibodies (eg, scFv), chimeric antibodies, diabody Diabody and a polypeptide comprising at least a portion of an antibody sufficient to confer specific ability to bind the antigen to the polypeptide.
- CDR complementarity determining region
- the term "Aptamer” means capable of high affinity and high specificity.
- G-tetramer (G) -tetramer) A thermodynamically stable three-dimensional structure that specifically binds to a target protein of interest or other biological target molecule by, for example, structural complementation, base stacking force, van der Waals force, hydrogen bonding, or electrostatic interaction.
- the aptamer may be DNA or RNA, and may also contain a nucleic acid analog (eg, a locked nucleic acid (LNA), a peptide nucleic acid (PNA), a glycol nucleic acid (GNA), or a threose nucleic acid (TNA).
- a nucleic acid analog eg, a locked nucleic acid (LNA), a peptide nucleic acid (PNA), a glycol nucleic acid (GNA), or a threose nucleic acid (TNA).
- LNA locked nucleic acid
- PNA peptide nucleic acid
- GNA glycol nucleic acid
- TAA threose nucleic acid
- targeting polypeptide refers to a polypeptide molecule that can specifically bind to a target protein of interest.
- the targeting polypeptide may comprise a natural amino acid, a synthetic amino acid or an amino acid mimetic that functions in a manner similar to a naturally occurring amino acid.
- Naturally occurring amino acids are those encoded by the genetic code and those amino acids that are later modified, for example, hydroxyproline, ⁇ -hydroxyglutamate, O-phosphoserine, phosphothreonine or phosphotyrosine.
- polypeptide solution it binds the target protein can be used to target affinity dissociation equilibrium constant (i.e., K D value) Describe.
- K D value affinity dissociation equilibrium constant
- K D value is greater than about 10 -3 M
- K D value is usually considered to represent a non-binding or non-specific binding.
- a targeting polypeptide that specifically binds to the target protein can be obtained by methods known to those skilled in the art, such as by phage display technology or protein microarray technology.
- the expression “response to IFN[alpha] treatment” refers to a condition in which a chronic hepatitis B patient develops a virological response or a serological response after treatment with IFN/PEG-IFN.
- the virological response means HBV DNA ⁇ 2000 IU/ml 6 months after treatment and 6 to 12 months after withdrawal;
- the serological response means HBeAg is negative and anti-HBe appears (see, EASL Clinical Practice Guidelines for Chronic Hepatitis B (2012 Edition)).
- the expression “non-responsive to IFN ⁇ treatment” refers to a condition in which a chronic hepatitis B patient does not satisfy the conditions described above after receiving IFN/PEG-IFN treatment.
- the term "statistical analysis value” refers to a value obtained by statistically analyzing the detection results obtained by various detection methods.
- a variety of statistical analysis methods are known in the art (see, for example, PCT International Application WO2009064901, which is incorporated herein by reference), and including but not limited to the linear combination of the test results, the linear regression model, the logistic regression model, the linear discriminant analysis ( LDA) model, nearest neighbor model or microarray predictive analysis (PAM).
- LDA linear discriminant analysis
- PAM microarray predictive analysis
- the statistical analysis value is a value obtained by statistical analysis by a logistic regression model.
- Logistic regression models are described in detail, for example, in Hu Chunyan. Four tumor markers are in Combined detection in ovarian cancer serum [D]. Guangzhou: Sun Yat-sen University, 2008: 1-39", which is incorporated herein by reference in its entirety.
- the term "reference value" refers to a value that reflects the condition of a population of CHB patients who are not responsive to IFN[alpha] treatment.
- the reference value includes, for example, a marker level in a CHB patient population sample according to a CHB patient population sample that is not responsive to IFN ⁇ treatment or a marker in a CHB patient population sample that does not respond to IFN ⁇ treatment before and after IFN ⁇ and/or HBV antigen stimulation.
- a level of change in the level (relative expression level), a normal value or range of values determined; and a test value obtained from a sample of a population of CHB patients who did not respond to treatment with IFN ⁇ (eg, the level of the marker described above) Change factor)
- a test value obtained from a sample of a population of CHB patients who did not respond to treatment with IFN ⁇ (eg, the level of the marker described above) Change factor)
- a value obtained by statistical analysis statistic analysis value.
- Methods for determining optimal diagnostic cutoff values are well known in the art and include, but are not limited to, Receiver Operating Characteristic (ROC) curve analysis, which is described in detail, for example, in "Habibzadeh F, et al., Biochem.
- Cycle threshold refers to the number of cycles experienced by a fluorescent signal within each reaction tube when it reaches a set threshold in a real-time PCR assay.
- Ct value of each template There is a linear relationship between the Ct value of each template and the logarithm of the initial copy number of the template. The more the starting copy number, the smaller the Ct value.
- a standard curve can be made using a standard of known starting copy number, where the logarithm of the starting copy number is plotted on the abscissa and the Ct value is taken as the ordinate. Therefore, as long as the Ct value of the unknown sample is obtained, the initial copy number of the sample can be calculated from the standard curve.
- the term "comparative Ct method” is a method of relative quantitative analysis of mRNA well known in the art, which is described in detail, for example, in Livak KJ, et al. Methods. 2001 Dec; 25(4): 402-8 ( It is incorporated herein by reference).
- quantitative PCR quantitative PCR
- each PCR cycle doubles the amplification product
- the Ct value obtained during the exponential phase of the PCR reaction can reflect the copy number of the starting template, so
- the difference in Ct values of different samples to be tested is one cycle, which means that their starting template copy number has a difference of 2 times. Based on this, by comparing the Ct values of different samples, the relative expression level of the target gene in the sample to be tested can be judged.
- normalization refers to the elimination of the possible variations in the various steps of the detection method employed by comparing the expression levels of the same gene in different samples. The difference in expression caused by the yield.
- Methods of normalization are known in the art, including but not limited to The method of PCT International Application No. WO2013068422A1 or Chinese Patent Application No. 201280035399.6, for example, a comparative Ct method based on an internal reference gene.
- the term "expression level" refers to a measurable amount of a gene product produced by a gene of interest in a sample from a subject, wherein the gene product can be a transcription product or a translation product.
- the expression "determining the expression level of a gene of interest” may mean determining the level of the mRNA or fragment of the mRNA of the gene, or the cDNA of the gene or the fragment of the cDNA, or is encoded by the gene. The level of the protein or its polypeptide fragment.
- PBMC Peripheral blood mononuclear cell
- T cells lymphocytes
- B cells lymphocytes
- NK cells monocytes or dendritic cells.
- Methods for obtaining PBMC from peripheral blood are well known in the art and include, but are not limited to, Ficoll stratified liquor or Percoll stratified liquor.
- reagent for isolating PBMC means an agent which is required in the method for obtaining PBMC described above, for example, a polysucrose-transfloxacin for Ficoll stratified liquid method. Amine solution.
- PBMC separation device means a device capable of separating PBMC from a sample (for example, peripheral blood) from a subject, such devices are known in the art including, but not limited to, detailed Cell separation devices are described in Chinese Patent Application Nos. CN1958776A, CN102286360A, and CN105132278A, all of which are incorporated herein by reference.
- peripheral blood leukocyte layer refers to a component formed by peripheral anticoagulation after natural sedimentation, centrifugation or density gradient centrifugation, mainly by white blood cells (including peripheral blood mononuclear cells) and Platelet composition. After anticoagulation, the upper layer of plasma, the lower layer of red blood cells, and a thin layer of white film between them, which accounts for about 1% of the total blood volume, are called the white film layer.
- the term "subject” includes, but is not limited to, various animals, with humans being particularly preferred.
- the term "diluent" is preferably an electrolyte solution capable of maintaining cell osmotic pressure, and if necessary, the solution also has a function of maintaining physiological pH.
- solutions are well known in the art and include, but are not limited to, Alsever's solution, Earle's Balanced Salt Solution (EBSS), Gey's Balanced Salt Solution (GBSS), Hanks' Balanced Salt Solution (HBSS), Phosphate Buffer (PBS), Dulbose Phosphate Buffer (DPBS), Puck's Balanced Salt Solution, Ringer's Balanced Salt Solution (RBSS), Simm's Balanced Salt Solution (SBSS), TRIS Buffer (TBS), Tyrode's Balanced Salt Solution (TBSS), Physiology Salt water or Ringer's Solution.
- the diluent is a phosphate buffer or physiological saline.
- anticoagulant refers to an agent or substance that is capable of preventing blood from clotting, such Substances are well known in the art and include, but are not limited to, heparin, EDTA, oxalates (e.g., sodium oxalate, potassium oxalate, ammonium oxalate), sodium citrate (sodium citrate).
- the term "culture fluid” or “medium” refers to a nutrient that is capable of maintaining cellular activity.
- the nutrients contain amino acids, vitamins, carbohydrates, inorganic salts and the like.
- Such nutrients are well known in the art and include, but are not limited to, RPMI-1640 medium or DMEM medium.
- the purpose of adding a culture solution or a medium to a sample from the subject is to maintain the activity of cells, particularly PBMC, in the sample.
- Methods for maintaining the activity of cells in blood components are well known in the art, and those skilled in the art can select according to actual needs.
- a culture solution when the sample is whole blood, a culture solution may be added, for example, adding a suitable amount of glucose, sodium chloride, potassium chloride, etc. in a phosphate buffer or physiological saline; In some embodiments, the culture solution is added with a suitable amount of glucose and potassium chloride in a phosphate buffer.
- a medium such as a cell culture medium, may be added, for example, suitable for maintaining blood cells, particularly PBMC. Active cell culture medium, such as RPMI-1640 medium or DMEM medium.
- the inventors of the present application after tracking the response of chronic hepatitis B patients to IFN ⁇ treatment, unexpectedly found that the level of OAS2 in the pre-treatment PBMC samples of patients treated with IFN ⁇ was significantly higher than that of patients who did not respond to IFN ⁇ treatment. Differences, or pre-treatment PBMC samples from patients treated with IFN ⁇ , showed significant differences in OAS2 and/or UPS18 levels compared to patients who did not respond to IFN ⁇ treatment after in vitro induction, so OAS2 and/or UPS18 could be used to predict CHB A marker of IFN ⁇ therapeutic response in patients.
- the field has long lacked a well-defined anti-HBV immune response index to predict the response of CHB patients to IFN ⁇ treatment. Based on this finding, the inventors have developed a new method for predicting the therapeutic effect of IFN ⁇ on CHB patients or the response of CHB patients to IFN ⁇ treatment.
- the invention provides a kit comprising a first reagent capable of detecting a level of expression of a marker, optionally further comprising an IFN alpha and/or a stimulator; wherein the marker is selected from OAS2 or In USP 18, the stimulator is selected from the group consisting of an HBV antigen, an antigenic fragment of an HBV antigen, or any combination thereof.
- the HBV antigen is HBcAg.
- the HBcAg has the amino acid sequence set forth in SEQ ID NO:38.
- the antigenic fragment has an amino acid sequence selected from the group consisting of SEQ ID NOs: 3-37.
- the stimuli comprise as shown in SEQ ID NOs: 3-37, respectively.
- the first agent is an agent capable of detecting the mRNA level of the marker.
- agents are well known in the art and include, but are not limited to, nucleic acid probes that specifically bind to a sequence of interest, primers that amplify a target sequence, non-specific fluorescent dyes (eg, SYBR Green I), or combinations thereof.
- the nucleic acid probe can be a single-labeled nucleic acid probe, such as a radionuclide (eg, 32 P, 3 H, 35 S, etc.) labeled probe, a biotinylated probe, or a horseradish.
- the nucleic acid probe may also be a double-labeled nucleic acid Probes, such as Taqman probes, molecular beacons, displacement probes, scorpion primer probes, QUAL probes, FRET probes, and the like.
- the first reagent comprises a Taqman probe.
- the cDNA of OAS2 has the nucleotide sequence set forth in SEQ ID NO: 1.
- the cDNA of USP18 has the nucleotide sequence set forth in SEQ ID NO:2.
- the first agent is an agent capable of detecting the protein level of the marker.
- agents are well known in the art and include, but are not limited to, antibodies, targeting polypeptides or nucleic acid aptamers that are capable of specifically binding to OAS2 or USP18 proteins.
- such agents carry a detectable label, such as an enzyme (eg, horseradish peroxidase, alkaline phosphatase, etc.), a radionuclide (eg, 3 H, 125 I, 35 S, 14) C, 32 P, etc.), fluorescent dyes (such as FITC, TRITC, PE, Texas Red, quantum dots, Cy7, Alexa 750, etc.), acridine esters, magnetic beads (for example, ), colloidal gold or colored glass or plastic (eg, polystyrene, polypropylene, latex, etc.) beads, and biotin for binding to the above-described label modified avidin (eg, streptavidin).
- an enzyme eg, horseradish peroxidase, alkaline phosphatase, etc.
- a radionuclide eg, 3 H, 125 I, 35 S, 14
- fluorescent dyes such as FITC, TRITC, PE, Texas Red, quantum dots, Cy7, Alex
- the first agent determines the protein level of OAS2 or USP18 in the sample by immunological detection.
- the immunological assay is selected from the group consisting of an ELISA assay, an Elispot assay, a Western blot, or a surface plasmon resonance method.
- the first agent comprises an antibody against an OAS2 or USP18 protein or an antigen binding fragment thereof.
- kits of the invention comprise a first agent capable of detecting the level of expression of OAS2, optionally further comprising IFN ⁇ and/or a stimulator.
- kits of the invention comprise a first agent and IFN[alpha] capable of detecting USP18 expression levels, optionally further comprising an irritant.
- the kit of the present invention further comprises a second reagent for pretreating a sample containing PBMC, wherein the second reagent comprises one or more reagents selected from the group consisting of: a diluent for the sample (eg, phosphate buffer or saline); an anticoagulant (eg, heparin) used to prevent blood clotting; used to maintain A reagent for PBMC activity (for example, a culture medium or a culture solution); and an agent for isolating PBMC (for example, a lymphocyte separation solution such as a sucrose-diammonia solution).
- a diluent for the sample eg, phosphate buffer or saline
- an anticoagulant eg, heparin
- a reagent for PBMC activity for example, a culture medium or a culture solution
- an agent for isolating PBMC for example, a lymphocyte separation solution such as a sucrose-diammonia solution.
- the second agent comprises an agent (eg, a culture medium or culture medium) for maintaining PBMC activity, and/or an agent for isolating PBMC (eg, a lymphocyte separation solution, eg, Polysucrose - diatrizoate solution).
- an agent eg, a culture medium or culture medium
- an agent for isolating PBMC eg, a lymphocyte separation solution, eg, Polysucrose - diatrizoate solution.
- the kit of the invention further comprises a blood collection device (eg, a pyrogen-free vacuum blood collection tube) and/or a PBMC separation device (eg, a PBMC cell separation tube).
- a blood collection device eg, a pyrogen-free vacuum blood collection tube
- a PBMC separation device eg, a PBMC cell separation tube
- the kit of the present invention further comprises an agent capable of detecting the expression level of the internal reference gene, the expression of the internal reference gene in each tissue and cell is relatively constant, and does not under the condition of the treatment factor Expression changes occur, such as a housekeeping gene; typically, the housekeeping gene encodes proteins necessary for maintaining essential cellular life activities including, but not limited to, ⁇ -actin, GAPDH, 18S rRNA, ⁇ 2-MG, UBC, or ⁇ -tubulin.
- the agent capable of detecting the expression level of an internal reference gene is an agent capable of detecting the mRNA level of the internal reference gene.
- the agent capable of detecting the expression level of an internal reference gene is an agent capable of detecting the protein level of the internal reference gene.
- the invention relates to the use of an agent capable of detecting the expression level of a marker for predicting the therapeutic effect of IFN ⁇ on a subject having chronic hepatitis B or the subject A response to IFN[alpha] treatment; wherein the marker is selected from the group consisting of OAS2 or USP18.
- the kit further comprises an IFN alpha and/or a stimulator selected from the group consisting of an HBV antigen, an antigenic fragment of an HBV antigen, or any combination thereof.
- the HBV antigen is HBcAg.
- the HBcAg has the amino acid sequence set forth in SEQ ID NO:38.
- the antigenic fragment has an amino acid sequence selected from the group consisting of SEQ ID NOs: 3-37.
- the stimulator comprises an antigenic fragment having an amino acid sequence as set forth in SEQ ID NOs: 3-37, respectively.
- the agent capable of detecting the level of expression of a marker is an agent capable of detecting the mRNA level of the marker.
- agents are well known in the art and include, but are not limited to, nucleic acid probes that specifically bind to a sequence of interest, primers that amplify a target sequence, non-specific fluorescent dyes (eg, SYBR Green I), or combinations thereof.
- the nucleic acid probe can be a single-labeled nucleic acid probe, such as a radionuclide (eg, 32 P, 3 H, 35 S, etc.) labeled probe, a biotinylated probe, or a horseradish.
- the nucleic acid probe may also be a double-labeled nucleic acid Probes, such as Taqman probes, molecular beacons, displacement probes, scorpion primer probes, QUAL probes, FRET probes, and the like.
- the agent capable of detecting the expression level of a marker comprises a Taqman probe.
- the cDNA of OAS2 has the nucleotide sequence set forth in SEQ ID NO: 1.
- the cDNA of USP18 has the nucleotide sequence set forth in SEQ ID NO:2.
- the mRNA level of the marker can be determined using mRNA detection methods known in the art, such as fluorescent quantitative PCR, Northern blotting, in situ hybridization, or including mRNA amplification (eg, reverse transcription PCR) and the mRNA amplification.
- a method of quantifying product eg, electrophoresis and staining.
- the agent capable of detecting the expression level of a marker quantifies the mRNA level of the marker by PCR.
- the agent capable of detecting the expression level of a marker determines the mRNA level of the marker by real-time PCR.
- the agent capable of detecting the level of expression of a marker is an agent capable of detecting the protein level of the marker.
- agents are well known in the art and include, but are not limited to, antibodies, targeting polypeptides or nucleic acid aptamers that are capable of specifically binding to OAS2 or USP18 proteins.
- such agents carry a detectable label, such as an enzyme (eg, horseradish peroxidase, alkaline phosphatase, etc.), a radionuclide (eg, 3 H, 125 I, 35 S, 14) C, 32 P, etc.), fluorescent dyes (such as FITC, TRITC, PE, Texas Red, quantum dots, Cy7, Alexa 750, etc.), acridine esters, magnetic beads (for example, ), colloidal gold or colored glass or plastic (eg, polystyrene, polypropylene, latex, etc.) beads, and biotin for binding to the above-described label modified avidin (eg, streptavidin).
- an enzyme eg, horseradish peroxidase, alkaline phosphatase, etc.
- a radionuclide eg, 3 H, 125 I, 35 S, 14
- fluorescent dyes such as FITC, TRITC, PE, Texas Red, quantum dots, Cy7, Alex
- the agent capable of detecting the expression level of a marker determines the protein level of OAS2 or USP18 in the sample by immunological detection.
- the immunological assay is selected from the group consisting of an ELISA assay, an Elispot assay, a Western blot, or a surface plasmon resonance method.
- the agent capable of detecting the expression level of a marker comprises an antibody against an OAS2 or USP18 protein or an antigen-binding fragment thereof.
- the kit further comprises an agent capable of detecting the expression level of the internal reference gene, the expression of the internal reference gene in each tissue and cell is relatively constant, and does not occur under the conditions of the treatment factor Expression alterations, such as housekeeping genes; typically, the housekeeping genes encode proteins necessary for maintaining essential life activities of the cell including, but not limited to, ⁇ -actin, GAPDH, 18S rRNA, ⁇ 2-MG, UBC or ⁇ -tubulin.
- the agent capable of detecting the level of expression of an internal reference gene is capable of detecting the internal reference group Reagents for mRNA levels.
- the agent capable of detecting the expression level of an internal reference gene is an agent capable of detecting the protein level of the internal reference gene.
- the kit further comprises one or more reagents or devices selected from 1) to 6):
- a diluent for diluting the sample such as phosphate buffer or physiological saline
- an anticoagulant for preventing blood coagulation such as heparin
- an agent for maintaining the activity of PBMC such as a medium or a culture solution
- an agent for isolating PBMC such as a lymphocyte separation solution, such as a sucrose-diammonia solution;
- a blood collection device such as a pyrogen-free vacuum blood collection tube
- PBMC separation device such as a PBMC cell separation tube.
- the kit includes an agent (eg, a culture medium or culture medium) for maintaining PBMC activity, and/or an agent for isolating PBMC (eg, a lymphocyte separation solution, such as a poly Sucrose - diatrizoate solution).
- an agent eg, a culture medium or culture medium
- an agent for isolating PBMC eg, a lymphocyte separation solution, such as a poly Sucrose - diatrizoate solution.
- the kit predicts the therapeutic effect of IFNa on a subject having chronic hepatitis B or the subject's response to IFN[alpha] treatment by a method comprising the steps of:
- the sample comprises peripheral blood mononuclear cells (PBMC), such as whole blood (eg, anticoagulated whole blood), peripheral blood mononuclear cells (PBMC), or a peripheral blood white layer.
- PBMC peripheral blood mononuclear cells
- the expression level when the expression level is greater than the reference value, or when the statistical analysis value of the expression level is greater than the reference value, indicating that the subject is responsive to IFN[alpha] treatment Suitably receiving IFN ⁇ treatment; when the expression level is not greater than the reference value, or when the statistical analysis value of the expression level is not greater than the reference value, indicating that the subject does not respond to IFN ⁇ treatment It is not suitable for IFN ⁇ treatment.
- step (2) the expression levels are statistically analyzed using a logistic regression model.
- the expression level of the marker to be tested in different samples can usually be used. Normalization is performed, for example, by comparing the expression level value of the marker to be tested in the sample with the expression level value of the reference gene.
- the expression level of the marker is a normalized expression level.
- the normalized expression level can be obtained by an exemplary method of determining the expression level of the internal reference gene in a sample from the subject using an agent capable of detecting the expression level of the internal reference gene, and expressing the marker The level is compared to the expression level of the reference gene to obtain a normalized expression level of the marker.
- the method further comprises one or more of the following steps: (a) obtaining a sample from the subject using a blood collection device; (b) using An anticoagulant treats a blood collection device or a sample from the subject; (c) treats a sample from the subject using an agent for maintaining PBMC activity; (d) dilutes from the subject using a diluent And (e) separating the PBMC from the sample from the subject using a reagent for separating PBMC or a PBMC separation device.
- the kit predicts the therapeutic effect of IFNa on a subject having chronic hepatitis B or the subject's response to IFN[alpha] treatment by a method comprising the steps of:
- the sample comprises peripheral blood mononuclear cells (PBMC), such as whole blood (eg, anticoagulated whole blood), peripheral blood mononuclear cells (PBMC), or a peripheral blood white layer.
- PBMC peripheral blood mononuclear cells
- the relative expression level when the relative expression level is less than the reference value, or when the statistical analysis value of the relative expression level is less than the reference value, indicating that the subject is to be treated with IFN ⁇ Generating a response suitable for IFN ⁇ treatment; when the relative expression level is not less than the reference value, or when the statistical analysis value of the relative expression level is not less than the reference value, indicating that the subject does not IFN ⁇ treatment produces a response that is not suitable for IFN ⁇ treatment.
- step (2) a change in the mRNA level of the marker in the test sample compared to the mRNA level of the marker in the control sample is obtained by comparing the Ct method. That is, the relative expression level of the mRNA of the marker in the sample to be tested.
- step (3) the relative expression levels are statistically analyzed using a logistic regression model.
- the expression level of the marker to be tested in different samples can usually be used. Normalization is performed, for example, by comparing the expression level value of the marker to be tested in the sample with the expression level value of the reference gene.
- the expression level of the marker is a normalized expression level.
- the normalized expression level can be obtained by an exemplary method of determining the expression level of the internal reference gene in a sample from the subject using an agent capable of detecting the expression level of the internal reference gene, and expressing the marker The level is compared to the expression level of the reference gene to obtain a normalized expression level of the marker.
- the expression level of the marker is a normalized mRNA level, which can be obtained by the following exemplary method: using the comparative Ct method to measure the marker in the sample to be tested The mRNA level of the analyte is compared to the mRNA level of the internal reference gene to obtain a normalized mRNA level of the marker. Further, a fold change of the normalized mRNA level of the marker in the sample to be tested compared to the normalized mRNA level of the marker in the control sample is obtained by comparing the Ct method, that is, the sample in the sample to be tested The relative expression level of the mRNA of the marker.
- the method further comprises one or more of the following steps: (a) obtaining a sample from the subject using a blood collection device; (b) using An anticoagulant treats a blood collection device or a sample from the subject; (c) treats a sample from the subject using an agent for maintaining PBMC activity; (d) dilutes from the subject using a diluent And (e) separating the PBMC from the sample from the subject using a reagent for separating PBMC or a PBMC separation device.
- the invention provides a method for predicting the therapeutic effect of IFN ⁇ on a subject having chronic hepatitis B Or a method of the subject's response to IFN ⁇ treatment, or a method of obtaining a result of a diagnostic assay, wherein the diagnostic assay is for predicting a therapeutic effect of IFN ⁇ on a subject having chronic hepatitis B or the subject
- the response to IFN ⁇ treatment includes the following steps:
- the sample comprises peripheral blood mononuclear cells (PBMC), such as whole blood (eg, anticoagulated whole blood), peripheral blood mononuclear cells (PBMC), or a peripheral blood white layer.
- PBMC peripheral blood mononuclear cells
- the method further comprises the step of administering to the subject IFN[alpha] to treat chronic hepatitis B, wherein the subject is judged to be IFN[alpha] therapeutically produced Answer.
- the expression level when the expression level is greater than the reference value, or when the statistical analysis value of the expression level is greater than the reference value, indicating that the subject is responsive to IFN[alpha] treatment Suitably receiving IFN ⁇ treatment; when the expression level is not greater than the reference value, or when the statistical analysis value of the expression level is not greater than the reference value, indicating that the subject does not respond to IFN ⁇ treatment It is not suitable for IFN ⁇ treatment.
- step (2) the mRNA level or protein level of the marker is determined.
- the mRNA level of the marker is determined.
- the mRNA level of the marker can be determined using mRNA detection methods known in the art, such as fluorescent quantitative PCR, Northern blotting, in situ hybridization, or including mRNA amplification (eg, reverse transcription PCR) and the mRNA amplification. A method of quantifying product (eg, electrophoresis and staining).
- the mRNA level of the marker is quantified by PCR.
- the mRNA level of the marker is determined by real-time PCR.
- the cDNA of OAS2 has the nucleotide sequence set forth in SEQ ID NO: 1.
- the cDNA of USP18 has the nucleotide sequence set forth in SEQ ID NO:2.
- the protein level of the marker is determined.
- the protein level of the marker is determined by immunological detection.
- the immunological assay is selected from the group consisting of an ELISA assay, an Elispot assay, a Western blot, or a surface plasmon resonance method.
- an antibody against OAS2 is used or Its antigen-binding fragment is used to detect the protein level of OAS2.
- step (3) the expression levels are statistically analyzed using a logistic regression model.
- the expression level of the marker to be tested in different samples can usually be used. Normalization is performed, for example, by comparing the expression level value of the marker to be tested in the sample with the expression level value of the reference gene.
- the expression level of the marker is a normalized expression level.
- the normalized expression level can be obtained by an exemplary method of determining the expression level of the internal reference gene in a sample from the subject using an agent capable of detecting the expression level of the internal reference gene, and expressing the marker The level is compared to the expression level of the reference gene to obtain a normalized expression level of the marker.
- step (1) prior to step (1), one or more of the following steps are further included: (a) obtaining a sample from the subject; (b) from the subject An anticoagulant such as heparin is added to the sample; (c) PBMC or a blood component containing PBMC (for example, a peripheral blood leukocyte layer) is obtained from a sample from the subject; (d) from the subject A culture medium or a medium is added to the sample of the tester; and, (e) the sample from the subject is diluted.
- an anticoagulant such as heparin is added to the sample
- PBMC or a blood component containing PBMC for example, a peripheral blood leukocyte layer
- the present invention provides a method for predicting a therapeutic effect of IFN ⁇ on a subject having chronic hepatitis B or a response of said subject to IFN ⁇ treatment, or a method of obtaining a result of a diagnostic assay, wherein The diagnostic assay is used to predict the therapeutic effect of IFNa on a subject with chronic hepatitis B or the subject's response to IFN[alpha] treatment, which includes the following steps:
- step (3) determining the expression level of the marker in each sample in the step (2), and obtaining a change ratio of the expression level of the marker in the sample to be tested compared to the expression level of the marker in the control sample, Using the change factor as a relative expression level of the marker in the sample to be tested, wherein the marker is selected from OAS2 or USP18;
- the sample comprises peripheral blood mononuclear cells (PBMC), such as whole blood (eg, anticoagulated whole blood), peripheral blood mononuclear cells (PBMC), or a peripheral blood white layer.
- PBMC peripheral blood mononuclear cells
- the method further comprises the step of administering to the subject IFN ⁇ to treat chronic hepatitis B, wherein the subject is judged to be IFN ⁇ therapeutically produced Answer.
- the relative expression level when the relative expression level is less than the reference value, or when the statistical analysis value of the relative expression level is less than the reference value, indicating that the subject is to be treated with IFN ⁇ Generating a response suitable for IFN ⁇ treatment; when the relative expression level is not less than the reference value, or when the statistical analysis value of the relative expression level is not less than the reference value, indicating that the subject does not IFN ⁇ treatment produces a response that is not suitable for IFN ⁇ treatment.
- the HBV antigen is HBVcAg.
- the HBVcAg has the amino acid sequence set forth in SEQ ID NO:38.
- the antigenic fragment has an amino acid sequence selected from the group consisting of SEQ ID NOs: 3-37.
- the stimulator comprises an antigenic fragment having an amino acid sequence as set forth in SEQ ID NOs: 3-37, respectively.
- the mRNA level of the marker is determined.
- the mRNA level of the marker can be determined using mRNA detection methods known in the art, such as fluorescent quantitative PCR, Northern blotting, in situ hybridization, or including mRNA amplification (eg, reverse transcription PCR) and the mRNA amplification. A method of quantifying product (eg, electrophoresis and staining).
- the mRNA level of the marker is quantified by PCR.
- the mRNA level of the marker is determined by real-time PCR.
- the cDNA of OAS2 has the nucleotide sequence set forth in SEQ ID NO: 1.
- the cDNA of USP18 has the nucleotide sequence set forth in SEQ ID NO:2.
- the protein level of the marker is determined. In some excellent In selected embodiments, in step (3), the protein level of the marker is determined by immunological detection. Further, in certain preferred embodiments, the immunological assay is selected from the group consisting of an ELISA assay, an Elispot assay, a Western blot, or a surface plasmon resonance method. In certain embodiments, in step (3), an antibody against the OAS2 or USP18 protein or antigen-binding fragment thereof is used to detect the protein level of the marker.
- step (3) a change in the mRNA level of the marker in the test sample compared to the mRNA level of the marker in the control sample is obtained by comparing the Ct method. That is, the relative expression level of the mRNA of the marker in the sample to be tested.
- step (4) the relative expression levels are statistically analyzed using a logistic regression model.
- the expression level of the marker to be tested in different samples can usually be used. Normalization is performed, for example, by comparing the expression level value of the marker to be tested in the sample with the expression level value of the reference gene.
- the expression level of the marker is a normalized expression level.
- the normalized expression level can be obtained by an exemplary method of determining the expression level of the internal reference gene in a sample from the subject using an agent capable of detecting the expression level of the internal reference gene, and expressing the marker The level is compared to the expression level of the reference gene to obtain a normalized expression level of the marker.
- the expression level of the marker is a normalized mRNA level, which can be obtained by the following exemplary method: using the comparative Ct method to measure the marker in the sample to be tested The mRNA level of the analyte is compared to the mRNA level of the internal reference gene to obtain a normalized mRNA level of the marker. Further, a fold change of the normalized mRNA level of the marker in the sample to be tested compared to the normalized mRNA level of the marker in the control sample is obtained by comparing the Ct method, that is, the sample in the sample to be tested The relative expression level of the mRNA of the marker.
- step (1) prior to step (1), one or more of the following steps are further included: (a) obtaining a sample from the subject; (b) from the subject An anticoagulant such as heparin is added to the sample; (c) PBMC or a blood component containing PBMC (for example, a peripheral blood leukocyte layer) is obtained from a sample from the subject; (d) from the subject A culture medium or a medium is added to the sample of the tester; and, (e) the sample from the subject is diluted.
- an anticoagulant such as heparin is added to the sample
- PBMC or a blood component containing PBMC for example, a peripheral blood leukocyte layer
- composition comprising a full length or a partial fragment of a HBV core antigen and/or IFN ⁇ .
- a marker for predicting the therapeutic effect of alpha interferon in a chronic hepatitis B patient wherein the marker is an Interferon-stimulated genes (ISGs).
- ISGs Interferon-stimulated genes
- a method of screening for a marker for predicting the therapeutic effect of alpha interferon in a chronic hepatitis B patient comprising the steps of:
- the difference in the level of ISGs mRNA in the IFN ⁇ treatment response group and the non-response group can be used as the basis for the therapeutic effect.
- a kit for predicting the therapeutic effect of alpha interferon in chronic hepatitis B patients comprising an HBV core antigen polypeptide segment and an IFN ⁇ inducer, and an agent for detecting markers OAS2 and USP18 levels.
- kits of item 6 wherein the HBV core antigen polypeptide segment is a polypeptide segment of amino acid 1-183 of the HBV core antigen.
- the present invention has found through a large number of experiments and repeated exploration that the expression levels of OAS2 and/or USP18 are significantly different between the two groups of CHB patients who respond to IFN ⁇ treatment and those who do not respond to IFN ⁇ treatment, thereby establishing a simple, A method for predicting the therapeutic effect of IFN ⁇ on a subject having chronic hepatitis B or the subject's response to IFN ⁇ treatment is convenient, rapid, and highly accurate and specific.
- Figure 1 shows the significant difference in OAS2 mRNA levels of PBMCs in CHB patients between the IFN ⁇ treatment response group (Rs) and the non-response group (NRs) under different treatment conditions.
- Figure 1a shows that PBMC is not induced in vitro, and the mRNA level of OAS2 in the response group is significantly higher than that in the non-responder group.
- Figure 1b shows that the relative expression level of mRNA of OAS2 in the response group was significantly lower than that in the non-responder group after PBMC was induced by IFN ⁇ in vitro. (Note: * indicates p ⁇ 0.05, **** indicates p ⁇ 0.0005)
- Figure 2 shows that there is a significant difference in USP18 mRNA levels of PBMCs in CHB patients between the IFN ⁇ treatment response group (Rs) and the non-response group (NRs) under different treatment conditions.
- Figure 2a shows that the relative expression level of USP18 mRNA in the response group was significantly lower than that in the non-response group after PBMC was induced by IFN ⁇ in vitro
- Figure 2b shows the relative expression level of USP18 mRNA in the response group after PBMC was induced by IFN ⁇ and HBcAg peptide libraries. Significantly lower than the no response group. (Note: material * indicates p ⁇ 0.001, material ** indicates p ⁇ 0.0005)
- Figure 3 shows the ROC curves for the OAS2N assay and the OAS2 (IFNa + N-N) assay. The results showed that the OAS2N assay and the OAS2 (IFNa+N-N) assay have good sensitivity and specificity for predicting the response of CHB patients to IFN ⁇ therapy.
- Figure 4 shows the ROC curve of the OAS2 (IFNa + N-N) assay, the USP18 (IFNa + N-N) assay, and a combination of both.
- the results showed that the OAS2 (IFNa+N-N) assay, the USP18 (IFNa+N-N) assay, and the combination of the two were very sensitive and specific for predicting the response of CHB patients to IFN ⁇ therapy.
- lymphocyte separation solution (Ficoll-page Tm PLUS, GE Helathcare Life Sciences) equivalent to whole blood was added.
- the diluted whole blood is slowly added to the lymphocyte separation liquid along the tube wall to maintain a layered state.
- the above PBMC cells were seeded in a 24-well plate, 5 x 10 5 cells were added to each well, and 1 ml of RPM1640 medium containing 10% fetal calf serum was added. A total of four wells are set for each specimen, wherein the first well is not added with an inducing substance (the inducing substance includes IFN ⁇ and HBcAg polypeptide library) labeled as N, and the second hole is added with IFN ⁇ (Andafen, Anhui Anke Bioengineering (Group) Co., Ltd.) (1 ⁇ 10 4 IU/mL), a peptide library containing 35 HBcAg antigenic fragments (SEQ ID NO: 3-37) (synthesized by Shanghai Bioengineering Co., Ltd.) (1 ⁇ g/) was added to the third well. Strips/mL, a total of 35), the fourth well was added to the above IFN ⁇ and HBcAg polypeptide libraries. After the culture plate was placed in a carbon dioxide incubator at
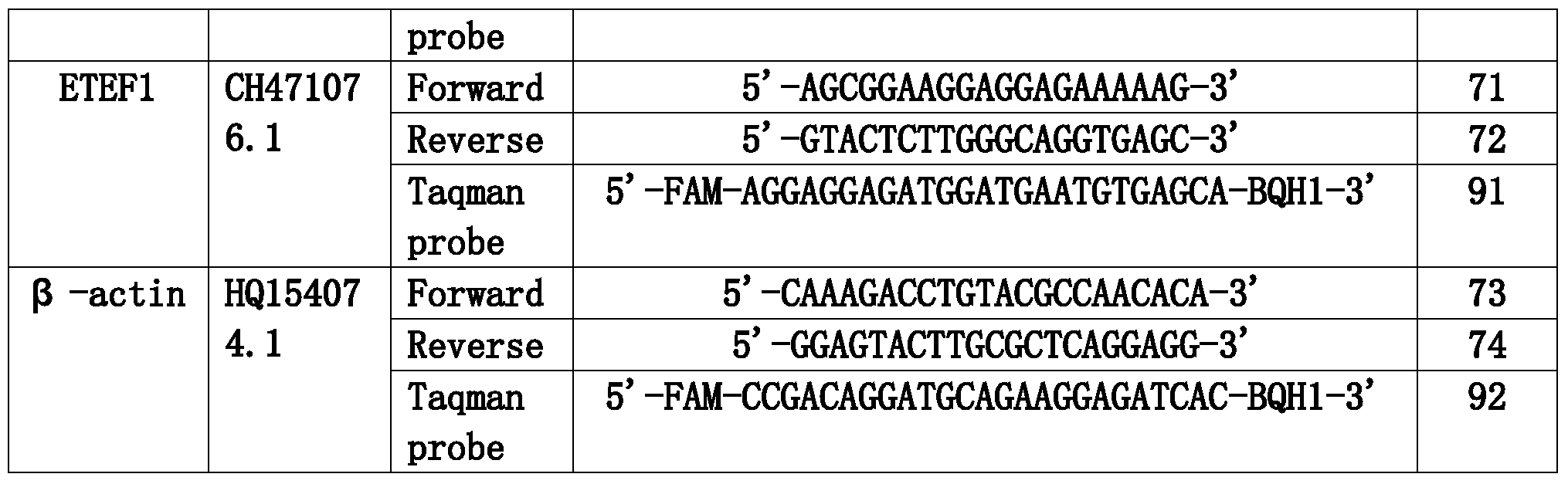
- the present invention has established fluorescence for 17 genes including the USP18, OAS2, LGP1, LAP3, GIP3, CEB1, ATF5, GIP2, OAS3, RPLP2, STXBP5, Viperin, RPS28, PI3KAP1, MX1, DUSP1, ETEF1 and the internal reference ⁇ -actin. Quantitative PCR detection system.
- Example 1 The PBMC cells obtained in Example 1 were collected by centrifugation, and the supernatant was discarded; 250 ⁇ l of RPMI1640 medium and 750 ⁇ l of Tripure were added to each well and mixed well; 200 ⁇ l of chloroform was added per 1 ml of cells, mixed upside down, and allowed to stand at 4 ° C. Set for 10min.
- RNA samples were determined using a spectrophotometer. 2 ⁇ L RNA sample was used to measure the absorbance of A260 and A280 in a 2 ⁇ L RNase-free water calibration instrument. The ratio of A260 to A280 was measured to evaluate the purity of the RNA sample. After detection, the RNA purity was A260/280 ⁇ 1.80.
- cDNA is a template in a fluorescent quantitative PCR detection system and stored at -20 °C.
- Pre-experiment was performed on 18 sets of primers designed by primer software (Table 2 above), and two templates from CHB patients (labeled CHB 1 and CHB 2, respectively) were randomly selected to detect the gene to be tested and the internal reference primer by real-time PCR. Ct value.
- the results of the identification are shown in Table 3.
- the Ct values of the primers of 17 sets of test genes and the ⁇ -actin primers of the internal reference were between 15-35, indicating that they could all be effectively amplified; while the Neg group (without template) was not detected.
- the 18 sets of primers listed in Table 2 above can be used for real-time PCR detection of the corresponding gene to be tested.
- the invention has systematically tracked 150 chronic hepatitis B specimens, among which 66 patients received IFN ⁇ treatment and 24 weeks of treatment results; among them, 47 patients had no response to IFN ⁇ treatment and 19 patients had IFN ⁇ treatment response.
- Table 4 shows The characteristic data of the selected study samples in this example showed that there was no significant difference in age and gender between the response group (Rs) and the non-response group (NRs).
- the results of ALT, AST and HBsAg, HBV DNA, and HBeAg before treatment were also There are no significant differences.
- Example 1.2 The specific steps of in vitro induction culture were carried out according to Example 1.2, wherein OAS2 (IFNa+NN) The group indicates the relative expression level of OAS2 mRNA in OAS2 relative to the control sample in the sample to be tested; USP18 (IFNa+NN) group and USP18 (IFNa+HBcAg-HBcAg) group respectively represent USP18 in the sample to be tested. Relative mRNA expression levels of USP18 relative to the respective control samples.
- Figure 1a shows the results of the OAS2N group. The results showed that the OAS2 normalized mRNA levels were significantly higher in the response group than in the non-responder group in the PBMC samples before the IFN ⁇ treatment without any in vitro induced CHB.
- Figure 1b shows the results of the OAS2 (IFNa+NN) group. The results showed that the relative expression level of OAS2 mRNA in the response group was significantly lower than that in the PBMC samples before the IFN ⁇ treatment. Group of patients.
- Figure 2a shows the results of the USP18 (IFNa+NN) group. The results showed that the relative expression level of USP18 mRNA in the response group was significantly lower than that in the PBMC samples before the IFN ⁇ treatment. Group of patients.
- Figure 2b shows the results of the USP18 (IFNa+HBcAg-HBcAg) group. The results showed that CHB patients receiving cultured in vitro via IFN ⁇ and HBcAg peptide libraries (containing HBcAg antigenic fragments shown in SEQ ID NO: 3-37) were accepted. In the PBMC samples before IFN ⁇ treatment, the relative expression level of USP18 mRNA in the response group was significantly lower than that in the non-responder group.
- OAS2 levels in pre-treatment PBMC samples in patients treated with IFN ⁇ are significantly higher than those in patients not responding to IFN ⁇ treatment; meanwhile, pre-treatment PBMC samples in IFN ⁇ -treated patients are induced in vitro by IFN ⁇ and/or HBcAg. Afterwards, the levels of OAS2 and/or UPS18 were significantly reduced compared to patients who did not respond to IFN[alpha] treatment. Thus OAS2 and/or UPS18 can be used to predict CHB patient response to IFN[alpha] treatment.
- Example 3 According to the experimental conditions of the OAS2N group and the OAS2 (IFNa+N-N) group in Example 3, the corresponding OAS2N detection method and OAS2 (IFNa+N-N) detection method were respectively established, and Table 6 shows the main steps of the above two methods.
- the CHB patient samples described in Example 3 were separately tested by the two methods described above, and two diagnostic indicators of the sample were obtained: OAS2 normalized mRNA level and OAS2 mRNA relative expression level.
- OAS2N assay or OAS2 (IFNa+NN) assay can be used to predict the therapeutic effect of IFN ⁇ on CHB patients or CHB patients on IFN ⁇ therapy. Response, with high sensitivity and specificity.
- Example 5 Analysis of OAS2 (IFNa+N-N) detection method, USP18 (IFNa+N-N) detection method and ROC curve analysis of combined detection
- the CHB patient samples described in Example 3 were respectively tested by the two methods described above, and two diagnostic indicators were obtained: the relative expression level of OAS2 mRNA and the relative expression level of USP18 mRNA;
- the relative expression level of mRNA of OAS2 or the relative expression level of mRNA of USP18 was used as a diagnostic index using SPSS 17.0 software, and the ROC curve was drawn to test the prediction of IFN ⁇ by OAS2 (IFNa+NN) assay or USP18 (IFNa+NN) assay alone.
Landscapes
- Life Sciences & Earth Sciences (AREA)
- Health & Medical Sciences (AREA)
- Chemical & Material Sciences (AREA)
- Engineering & Computer Science (AREA)
- Organic Chemistry (AREA)
- Genetics & Genomics (AREA)
- Zoology (AREA)
- Wood Science & Technology (AREA)
- Bioinformatics & Cheminformatics (AREA)
- Molecular Biology (AREA)
- General Engineering & Computer Science (AREA)
- Biotechnology (AREA)
- Proteomics, Peptides & Aminoacids (AREA)
- Biochemistry (AREA)
- Physics & Mathematics (AREA)
- Microbiology (AREA)
- Immunology (AREA)
- Biophysics (AREA)
- General Health & Medical Sciences (AREA)
- Biomedical Technology (AREA)
- Analytical Chemistry (AREA)
- Virology (AREA)
- Communicable Diseases (AREA)
- Plant Pathology (AREA)
- Measuring Or Testing Involving Enzymes Or Micro-Organisms (AREA)
Abstract
本发明提供了用于预测慢性乙肝患者对IFNα治疗的应答的标志物。本发明还提供了用于预测慢性乙肝患者对IFNα治疗的应答的方法,其包括测定慢性乙肝患者PBMC中这类标志物表达水平的步骤。本发明还提供了用于上述方法的试剂盒。
Description
本发明涉及用于预测慢性乙肝患者对IFNα治疗的应答的标志物。本发明还涉及用于预测慢性乙肝患者对IFNα治疗的应答的方法,其包括测定慢性乙肝患者PBMC中这类标志物表达水平的步骤。本发明还涉及用于上述方法的试剂盒。
乙型肝炎病毒感染,尤其是慢性乙型肝炎病毒感染是全球最为重要的公共卫生问题之一,目前全球约有超过3.5亿的慢性乙型肝炎病毒感染者。慢性乙型肝炎病毒感染可造成慢性乙型病毒性肝炎(Chronic hepatitis B,CHB)、肝硬化(Liver cirrhosis,LC)和原发性肝细胞癌(Hepatocellular carcinoma,HCC)等肝脏疾病,由慢性乙型肝炎病毒感染及其所引起的相关疾病所导致的死亡,全球每年超过100万人。
大部分的慢性HBV感染患者表现为无明显临床症状,其中约10-30%的患者发展成肝硬化或者肝癌。慢性HBV感染是一个动态过程,其感染的自然史可划分为4个不同的、且不一定存在必然联系的阶段:1)免疫耐受阶段,特点是HBeAg呈阳性,血清HBV DNA复制活跃,ALT水平正常或较低,一般不发生纤维化;2)主动免疫阶段/免疫清除解阶段,特点是HBeAg呈阳性但其水平开始下降,HBV DNA复制水平降低,血清HBsAg水平下降,ALT持续升高或波动。处于此期患者的CTL反应被激活,容易出现持续时间不等的中度或严重的炎症反应和肝纤维化;3)非活动携带阶段,这个阶段患者HBV DNA处于低复制期,DNA水平极低甚至检测不出来,HBsAg水平显著降低,ALT水平显示正常,肝组织没有或仅有轻度炎症,HBeAg阴性、anti-HBe阳性,这个时期意味着患者的HBV感染已得到免疫控制,也预示着一个长期且较好的预后,大部分患者发生肝硬化甚至肝癌的几率很低;4)HBeAg阴性肝炎期,属于慢性乙肝的晚期阶段,其主要特点为HBV DNA和ALT水平周期性波动以及活动性肝炎,并以前C区和/或基本核心启动子(BCP)变异的HBV病毒为主,不能表达或表达极低水平的HBeAg。
HBV病毒对宿主细胞并没有直接的致病作用,其主要的致病机理是HBV病毒引起宿主免疫应答,免疫应答进而引起免疫肝组织损伤。慢性HBV感染的治疗方式需要根据患者个体的不同情况来给予不同的抗病毒治疗。抗病毒治疗的目的是使HBV病毒持续被抑制,
阻止肝炎向纤维化、肝硬化甚至是肝癌的发展或转化,延长患者的生存期及提高生活质量。
目前有8种药物可用于CHB抗病毒治疗,包括6种核苷类似物:拉米夫定(LAM)、替比夫定(LdT)、恩曲他滨(FTC)、恩替卡韦(ETV)、阿德福韦(ADV)、替诺福韦(TDF);2种干扰素:普通干扰素(IFNα)和聚乙二醇干扰素(PEG-IFNα)。核苷类似物的主要作用是通过抑制HBV聚合酶的活性来抑制HBV病毒的复制。1976年Greenberg首次报道了IFNα治疗慢性乙型肝炎的疗效,IFNα是最早被FDA批准用于治疗慢性乙肝的药物,PEG-IFNα也被作为治疗慢性乙肝的首选药物,具有抗病毒和免疫调节的双重作用。IFNα的主要优点是无耐药性、并且具有免疫介导的控制HBV感染的作用,从而使治疗结束时患者有更高机会出现HBeAg血清学转换、更持久的病毒学应答以及HBsAg清除,使患者中的HBV DNA水平维持在检测下限。
抗病毒治疗必须保证一定程度上的病毒学抑制并能够产生生化缓解,组织学恢复及防止并发症的发生。理想的HBV治疗终点是HBsAg消失,但是现有的抗病毒药物难以达到这个目标,所以更现实的治疗终点是能够诱导一个持续的病毒学抑制状态。抗病毒应答可以分为生化水平、血清水平、病毒学水平和组织学水平。所有的应答都可以在治疗过程中和治疗后的某些时间点来进行评估。临床研究显示,CHB患者使用IFNα常规治疗6个月的应答率仅为25-40%。测定CHB患者针对HBV特异性免疫应答能力可能能够预测CHB患者接受治疗的预期疗效。长期以来,由于缺乏意义明确的抗HBV免疫应答指标,CHB患者血清ALT水平被作为衡量宿主抗HBV免疫能力的间接替代指标,但预测结果并不理想。若能在治疗前进行治疗应答预测,对于优化药物选择,提高治疗依从性以及保障疗效具有十分重要的意义。
因此,本领域需要能够简单、方便、快速且具有高准确性和特异性地预测CHB患者对IFNα治疗的应答的标志物。
发明内容
在本发明中,除非另有说明,否则本文中使用的科学和技术名词具有本领域技术人员所通常理解的含义。并且,本文中所用的细胞培养、生物化学、核酸化学、免疫学实验室操作步骤均为相应领域内广泛使用的常规步骤。同时,为了更好地理解本发明,下面提供相关术语的定义和解释。
如本文中所使用的,术语“IFNα”或“α型干扰素”包括所有天然或重组的α型干扰素,特别优选地为人α型干扰素,例如重组人α型干扰素,包括但不限于IFNα-1b(例如可获得自Schering Corporation,Kenilworth,N.J.的干扰素)、IFNα-2a(例如可获得自Hoffmann-La Roche,Nutley,N.J.的干扰素)或IFNα-2b(例如可获得自Schering Corporation,Kenilworth,N.J.的干扰素);例如天然α型干扰素的混合物,包括但不限于IFNα-n1(例如可获得自Sumitomo,Japan的或Glaxo-Wellcome Ltd.,London,Great Britain的干扰素α-n1)或IFNα-n3(例如可获得自Interferon Sciences的Alferon干扰素)。在本发明中,术语“IFNα”或“α型干扰素”还包括任何具有IFNα生物学活性的物质,例如突变或修饰过的IFNα,例如IFNα的PEG衍生物(PEG-IFNα)。在本发明中,术语“IFNα”或“α型干扰素”不受任何特定的获得来源的限制,可通过市售来源获得或通过本领域技术人员已知的常规技术产生,所述生产方法包括但不限于生物来源提取法和基因工程提取法,其详细描述于例如“Pestka S.Arch Biochem Biophys.1983Feb15;221(1):1-37”(其通过引用并入本文)。
如本文中所使用的,术语“HBV抗原”是指,存在于乙型肝炎病毒(HBV)中能够诱发机体产生免疫反应的蛋白,包括乙型肝炎病毒核心抗原(HBcAg)、乙型肝炎病毒表面抗原(HBsAg)和乙型肝炎病毒E抗原(HBeAg)。
如本文中所使用的,术语“HBcAg”是指,乙型肝炎病毒(HBV)的核心抗原蛋白,其是本领域技术人员公知的(参见,例如GENBANK登录号:CAM31905.1)。
在本发明中,当提及HBcAg的氨基酸序列时,参照SEQ ID NO:38所示的序列来进行描述。然而,本领域技术人员理解,在HBcAg的氨基酸序列中,可天然产生或人工引入突变或变异(包括但不限于,置换,缺失和/或添加,例如不同基因型、基因亚型或不同血清型、血清亚型的HBcAg),而不影响其生物学功能。因此,在本发明中,术语“HBcAg”应包括所有此类序列,包括例如SEQ ID NO:38所示的序列以及其天然或人工的变体。并且,当描述HBcAg的序列片段时,其不仅包括SEQ ID NO:38的序列片段,还包括其天然或人工变体中的相应序列片段。例如,表述“HBcAg的第1-183位氨基酸残基”包括,SEQ ID NO:38的第1-183位氨基酸残基,以及其变体(天然或人工)中的相应片段。在本发明中,表述“相应片段”是指,当对序列进行最优比对时,即当序列进行比对以获得最高百分数同一性时,进行比较的序列中位于等同位置的片段。
在本发明中,术语“HBcAg”不受任何特定的合成蛋白的方法限制,可通过本领域技术人员已知的常规技术产生,例如DNA重组技术或化学合成技术。
如本文中所使用的,表述“HBcAg的抗原性片段”是指,HBcAg蛋白经过截短后得到的氨基酸序列片段(即,多肽),该片段具有与相应的全长蛋白相同的生物学活性,即,可刺激并活化来自慢性乙肝患者的PBMC。例如本发明中SEQ ID NO:3-37所示的氨基酸序列片段即为HBcAg的抗原性片段。在本发明中,该抗原性片段不受任何特定的合成多肽的方法限制,可通过本领域技术人员已知的常规技术产生,例如DNA重组技术或化学合成技术。
在本发明中,HBcAg或其抗原性片段(例如,分别具有如SEQ ID NO:3-37所示的氨基酸序列的抗原性片段)可以通过DNA重组技术获得,例如通过使用无细胞表达系统从编码这些蛋白或多肽的多核苷酸获得(无细胞表达系统包括例如基于网织红细胞裂解物的表达系统、基于麦胚提取物的表达系统以及基于大肠杆菌提取物的表达系统);或通过使用体内表达系统(例如,大肠杆菌原核表达系统、酵母真核表达系统)从编码这些蛋白或多肽的多核苷酸获得。作为另外一种选择,HBcAg或其抗原性片段(例如,分别具有如SEQ ID NO:3-37所示的氨基酸序列的抗原性片段)可以通过化学合成产生。蛋白或多肽化学全合成的方法在本领域内是熟知的(参见,例如,RaibautL,et al.,Top Curr Chem.2015;363:103-54;Thapa P,et al.Molecules.2014;19(9):14461-83;Dawson PE,et al.,Science,1994;266(5186):776-9;和Wang P,et al.,Tetrahedron Lett,1998,39(47):88711-14;其通过引用并入本文),并且包括但不限于:固相肽合成技术(Solid Phase Peptide Synthesis,SPPS)或液相分段合成技术(例如,天然化学连接法(Native Chemical Ligation,NCL)、叠氮法(Azide method)、转移活化酯法(Transfer Active Ester Condensation,TAEC))。
如本文中所使用的,表述“分别具有如SEQ ID NO:3-37所示的氨基酸序列的抗原性片段”是指,具有SEQ ID NO:3所示的氨基酸序列的抗原性片段、具有SEQ ID NO:4所示的氨基酸序列的抗原性片段、…具有SEQ ID NO:36所示的氨基酸序列的抗原性片段、和具有SEQ ID NO:37所示的氨基酸序列的抗原性片段的组合。
如本文中所使用的,术语“免疫学检测”是指,利用抗原-抗体之间的特异性相互作用/结合亲和力来进行的测定,其一般可用于检测特定抗原或者抗体在样品中的存在或水平。此类免疫学测定是本领域技术人员公知的,包括但不限于,ELISA检测,Elispot检
测,Western印迹,表面等离子共振法等。关于免疫学测定的详细描述,可参见例如,Fundamental Immunology,Ch.7Paul,W.,ed.,第2版,Raven Press,N.Y.(1989)。
如本文中所使用的,术语“抗体”是指,通常由两对多肽链(每对具有一条“轻”(L)链和一条“重”(H)链)组成的免疫球蛋白分子。抗体轻链可分类为K和λ轻链。重链可分类为μ、δ、γ、α或ε,并且分别将抗体的同种型定义为IgM、IgD、IgG、IgA和IgE。在轻链和重链内,可变区和恒定区通过大约12或更多个氨基酸的“J”区连接,重链还包含大约3个或更多个氨基酸的“D”区。各重链由重链可变区(VH)和重链恒定区(CH)组成。重链恒定区由3个结构域(CH1、CH2和CH3)组成。各轻链由轻链可变区(VL)和轻链恒定区(CL)组成。轻链恒定区由一个结构域CL组成。抗体的恒定区可介导免疫球蛋白与宿主组织或因子,包括免疫系统的各种细胞(例如,效应细胞)和经典补体系统的第一组分(Clq)的结合。VH和VL区还可被细分为具有高变性的区域(称为互补决定区(CDR)),其间散布有较保守的称为构架区(FR)的区域。各VH和VL由按下列顺序:FR1、CDR1、FR2、CDR2、FR3、CDR3、FR4从氨基末端至羧基末端排列的3个CDR和4个FR组成。各重链/轻链对的可变区(VH和VL)分别形成抗体结合部位。氨基酸至各区域或结构域的分配遵循Kabat Sequences of Proteins of Immunological Interest(National Institutes of Health,Bethesda,Md.(1987and 1991)),或Chothia&Lesk(1987)J.Mol.Biol.196:901-917;Chothia等人(1989)Nature 342:878-883的定义。术语“抗体”不受任何特定的产生抗体的方法限制。例如,其包括,特别地,重组抗体、单克隆抗体和多克隆抗体。抗体可以是不同同种型的抗体,例如,IgG(例如,IgG1,IgG2,IgG3或IgG4亚型),IgA1,IgA2,IgD,IgE或IgM抗体。
如本文中所使用的,抗体的“抗原结合片段”是指,全长抗体的一个或多个部分,所述部分保持结合抗体所结合的相同抗原(例如,OAS2或USP18)的能力,能够与完整抗体竞争对抗原的特异性结合。通常参见,Fundamental Immunology,Ch.7Paul,W.,ed.,第2版,Raven Press,N.Y.(1989),其以其全文通过引用合并入本文,用于所有目的。可通过重组DNA技术或通过完整抗体的酶促或化学断裂产生抗原结合片段。在一些情况下,抗原结合片段包括Fab、Fab’、F(ab’)2、Fd、Fv、dAb和互补决定区(CDR)片段、单链抗体(例如,scFv)、嵌合抗体、双抗体(diabody)和这样的多肽,其包含足以赋予多肽特异性结合抗原能力的抗体的至少一部分。
如本文中所使用的,术语“核酸适体(Aptamer)”是指,能够高亲和性和高特异性
地结合目的靶蛋白或其它生物靶分子的单链寡核苷酸,其可折叠形成例如茎环(Stem-Loop)、发夹(Hairpin)、假结(Pseudoknot)或G-四聚体(G-tetramer)的热力学稳定的三维空间结构,通过例如结构互补、碱基堆积力、范德华力、氢键或静电作用与目的靶蛋白或其它生物靶分子特异性结合。核酸适体可以为DNA或RNA,也可以包含核酸类似物(例如锁核酸(LNA)、肽核酸(PNA)、二醇核酸(GNA)或苏糖核酸(TNA))。获得结合特定靶蛋白的核酸适体的方法是本领域公知的,例如SELEX(Systematic evolution of ligands by exponential enrichment)筛选技术。
如本文中所使用的,术语“靶向多肽”是指,可以特异性结合目的靶蛋白的多肽分子。在本发明中,该靶向多肽可包含天然氨基酸、合成的氨基酸或采用与天然存在的氨基酸类似的方式起作用的氨基酸模拟物(mimetics)。天然存在的氨基酸为通过遗传密码来编码的那些以及后来修饰的那些氨基酸,例如,羟基脯氨酸、γ-羟基谷氨酸盐、O-磷酸丝氨酸、磷酸苏氨酸或磷酸酪氨酸。在本发明中,可基于亲和力来确定靶向多肽与其目的靶蛋白之间的“特异性”,该亲和力可用靶向多肽与其所结合的目的靶蛋白的解离平衡常数(即,KD值)进行描述。KD值越低,靶向多肽与其所结合的目的靶蛋白之间的结合强度越强。在本领域中通常已知,大于约10-3M的KD值通常被认为表示非结合或非特异性结合。取决于具体的目的靶蛋白,可以通过本领域技术人员已知的方法获得特异性结合该靶蛋白的靶向多肽,例如通过噬菌体展示技术或蛋白质微阵列技术进行筛选。
如本文中所使用的,表述“对IFNα治疗应答”是指,慢性乙肝患者接受IFN/PEG-IFN治疗后,出现病毒学应答或血清学应答的情况。其中,所述病毒学应答是指治疗后6个月HBV DNA<2000IU/ml,并且停药后维持6到12个月;所述血清学应答是指HBeAg阴转并出现anti-HBe(参见,EASL慢性乙型肝炎临床实践指南(2012版))。相应地,表述“对IFNα治疗无应答”是指,慢性乙肝患者接受IFN/PEG-IFN治疗后,未满足上文所描述的条件的情况。
如本文中所使用的,术语“统计分析值”是指,对通过各种检测方法得到的检测结果进行统计分析后得到的值。各种统计分析方法是本领域公知的(参见,例如PCT国际申请WO2009064901,其通过引用并入本文),并且包括但不限于检测结果的线性组合、线性回归模型、Logistic回归模型、线性判别分析(LDA)模型、最近邻模型或微阵列预测分析(PAM)。通常而言,特别优选的是,所述统计分析值为通过Logistic回归模型进行统计分析得到的值。Logistic回归模型具体描述于例如“胡春艳.四种肿瘤标志物在
卵巢癌血清中的联合检测[D].广州:中山大学,2008:1-39”,其全部通过引用并入本文。
如本文中所使用的,术语“参考值”(也称为最佳诊断界值)是指能够反映对IFNα治疗无应答的CHB患者群体的状况的值。在本发明中,参考值包括例如,根据对IFNα治疗无应答的CHB患者群体样品中的标志物水平或根据IFNα和/或HBV抗原刺激前后对IFNα治疗无应答的CHB患者群体样品中的标志物水平的变化倍数(相对表达水平),所确定的一个正常数值或数值范围;以及,对从对IFNα治疗无应答的CHB患者群体的样品获得的检测值(例如,前文所述的标志物水平的变化倍数)进行统计分析而得到的值(统计分析值)。确定最佳诊断界值的方法是本领域熟知的,包括但不限于受试者工作特征曲线分析(Receiver Operating Characteristic(ROC)curve analysis),其详细描述于例如“Habibzadeh F,et al.,Biochem Med(Zagreb).2016;26(3):297-307”和“陈卫中等.ROC曲线中最佳工作点的选择[J].中国卫生统计,2006,23:157-158”,其全部通过引用并入本文。
如本文中所使用的,术语“Ct值(Cycle threshold,循环阈值)”是指,在荧光定量PCR检测中,每个反应管内的荧光信号到达设定阈值时所经历的循环数。每个模板的Ct值与该模板的起始拷贝数的对数存在线性关系,起始拷贝数越多,Ct值越小。利用已知起始拷贝数的标准品可作出标准曲线,其中以起始拷贝数的对数为横坐标,以Ct值作为纵坐标。因此,只要获得未知样品的Ct值,即可从标准曲线上计算出该样品的起始拷贝数。
如本文中所使用的,术语“比较Ct法”是本领域熟知的一种mRNA相对定量分析方法,其详细描述于例如Livak KJ,等人.Methods.2001Dec;25(4):402-8(其通过引用并入本文)。在进行实时PCR(定量PCR)反应过程中,可以假设每个PCR循环增加一倍的扩增产物,而在PCR反应的指数期得出的Ct值可以反映出起始模板的拷贝数,因此如果不同待测样品的Ct值相差一个循环,则意味着它们的起始模板拷贝数具有2倍的差异。基于此,通过比较不同样品的Ct值,即可对待测样品中目的基因的相对表达水平作出判断。
如本文中所使用的,术语“归一化(normalization)”或“标准化”是指,通过比较不同样品中同一个基因的表达水平以消除由所采用的检测方法各个步骤中可能存在的可变产率所导致的表达差异。归一化的方法是本领域已知的,包括但不限于详细描述于
PCT国际申请WO2013068422A1或中国专利申请201280035399.6中的方法,例如基于内参基因的比较Ct法。
如本文中所使用的,术语“表达水平”是指,来自受试者的样品中由目的基因产生的基因产物的可测量的量,其中基因产物可以是转录产物或翻译产物。因此,表述“测定目的基因的表达水平”可以指,测定所述基因的mRNA或所述mRNA的片段的水平、或所述基因的cDNA或所述cDNA的片段的水平、或由所述基因编码的蛋白或其多肽片段的水平。
如本文中所使用的,术语“外周血单个核细胞(Peripheral blood mononuclear cell,PBMC)”是指,外周血中具有单个细胞核的细胞的总称,包括但不限于淋巴细胞(T细胞、B细胞、NK细胞)、单核细胞或树突状细胞。从外周血中获得PBMC的方法是本领域熟知的,包括但不限于Ficoll分层液法或Percoll分层液法。在本发明中,术语“用于分离PBMC的试剂”是指,在上文所描述的获得PBMC的方法中所需要用到的试剂,例如用于Ficoll分层液法的聚蔗糖-泛影葡胺溶液。在本发明中,术语“PBMC分离装置”是指,能够从来自受试者的样品(例如,外周血)中分离获得PBMC的装置,这类装置是本领域已知的,包括但不限于详细描述于中国专利申请CN1958776A、CN102286360A以及CN105132278A中的细胞分离装置,其全部通过引用并入本文。
如本文中所使用的,术语“外周血白膜层”是指,外周抗凝血经过自然沉降、离心或密度梯度离心后形成的一个组分,主要由白细胞(包括外周血单个核细胞)和血小板组成。抗凝血经过离心以后会形成上层的血浆、下层的红细胞以及二者之间薄薄的一层白色膜状物,约占血液总体积的1%,被称为白膜层。
如本文中所使用的,术语“受试者”包括但不限于各种动物,特别优选的是人。
在本发明中,术语“稀释剂”优选为能够维持细胞渗透压的电解质溶液,必要时该溶液也具备保持生理pH值的作用。这类溶液是本领域熟知的,包括但不限于阿氏液(Alsever’s solution)、Earle’s平衡盐溶液(EBSS)、Gey’s平衡盐溶液(GBSS)、Hanks’平衡盐溶液(HBSS)、磷酸盐缓冲液(PBS)、杜氏磷酸盐缓冲液(DPBS)、Puck’s平衡盐溶液、Ringer’s平衡盐溶液(RBSS)、Simm’s平衡盐溶液(SBSS)、TRIS缓冲液(TBS)、Tyrode’s平衡盐溶液(TBSS)、生理盐水或林格氏液(Ringer’s Solution)。在某些实施方式中,所述稀释剂为磷酸盐缓冲液或生理盐水。
如本文中所使用的,术语“抗凝剂”是指,能够阻止血液凝固的试剂或物质,这类
物质是本领域熟知的,包括但不限于肝素、EDTA、草酸盐(例如,草酸钠、草酸钾、草酸铵)、枸橼酸钠(柠檬酸钠)。
如本文中所使用的,术语“培养液”或“培养基”是指,能够维持细胞活性的养料。通常所述养料含有氨基酸、维生素、碳水化合物、无机盐等。这类养料是本领域熟知的,包括但不限于RPMI-1640培养基或DMEM培养基。在本发明中,在来自所述受试者的样品中加入培养液或培养基的目的是为了维持样品中的细胞特别是PBMC的活性。维持血液成分中细胞的活性的方法为本领域所公知,本领域技术人员可以根据实际需要进行选择。在某些实施方案中,当样品为全血时,可以加入培养液,所述培养液例如为在磷酸盐缓冲液或生理盐水中添加适量的葡萄糖、氯化钠和氯化钾等;在某些实施方案中,所述培养液为在磷酸盐缓冲液中添加适量的葡萄糖和氯化钾。在某些实施方案中,当样品为外周血单个核细胞(PBMC)、外周血白膜层或其它含有PBMC的血液成分时,可以加入培养基,例如细胞培养基,例如适合维持血细胞特别是PBMC活性的细胞培养基,例如RPMI-1640培养基或DMEM培养基。
本申请的发明人在对慢性乙肝患者对IFNα治疗的应答情况进行追踪研究后,出人意料地发现,对IFNα治疗应答患者的治疗前PBMC样本中OAS2水平与对IFNα治疗无应答患者相比表现出明显差异,或对IFNα治疗应答患者的治疗前PBMC样本经体外诱导后其中OAS2和/或UPS18水平与对IFNα治疗无应答患者相比表现出明显差异,因而OAS2和/或UPS18可以作为用于预测CHB患者IFNα治疗应答的标志物。而在本申请之前,本领域长期以来缺乏意义明确的抗HBV免疫应答指标以预测CHB患者对IFNα治疗的应答情况。基于这一发现,本发明人开发了新的用于预测IFNα对CHB患者的治疗效果或CHB患者对IFNα治疗的应答的方法。
因此,在一个方面,本发明提供了一种试剂盒,其包括能够检测标志物表达水平的第一试剂,任选地还包括IFNα和/或刺激物;其中,所述标志物选自OAS2或USP18,所述刺激物选自HBV抗原、HBV抗原的抗原性片段或其任何组合。
在某些优选的实施方案中,所述HBV抗原为HBcAg。在某些优选的实施方案中,所述HBcAg具有如SEQ ID NO:38所示的氨基酸序列。在某些优选的实施方案中,所述抗原性片段具有选自下列的氨基酸序列:SEQ ID NO:3-37。
在一个特别优选的实施方案中,所述刺激物包含分别具有如SEQ ID NO:3-37所示
的氨基酸序列的抗原性片段。
在某些优选的实施方案中,所述第一试剂为能够检测所述标志物的mRNA水平的试剂。这类试剂是本领域公知的,包括但不限于特异性结合目标序列的核酸探针、扩增目标序列的引物、非特异性荧光染料(例如,SYBR Green I)或其组合。在某些实施方式中,所述核酸探针可以为单标记的核酸探针,例如放射性核素(如32P、3H、35S等)标记探针、生物素标记探针、辣根过氧化物酶标记探针、地高辛标记探针或荧光基团(如FITC、FAM、TET、HEX、TAMRA、Cy3、Cy5等)标记探针;所述核酸探针也可以为双标记的核酸探针,例如Taqman探针、分子信标、置换探针、蝎子引物探针、QUAL探针、FRET探针等。在某些实施方式中,所述第一试剂包括Taqman探针。在某些实施方式中,所述OAS2的cDNA具有如SEQ ID NO:1所示的核苷酸序列。在某些实施方式中,所述USP18的cDNA具有如SEQ ID NO:2所示的核苷酸序列。
在某些优选的实施方案中,所述第一试剂为能够检测所述标志物的蛋白水平的试剂。这类试剂是本领域熟知的,包括但不限于能够和OAS2或USP18蛋白特异性结合的抗体、靶向多肽或核酸适体。在某些实施方式中,这类试剂带有可检测的标记,例如酶(如辣根过氧化物酶、碱性磷酸酶等)、放射性核素(如3H、125I、35S、14C、32P等)、荧光染料(如FITC、TRITC、PE、Texas Red、量子点、Cy7、Alexa 750等)、吖啶酯类化合物、磁珠(例如,)、胶体金或有色玻璃或塑料(例如,聚苯乙烯、聚丙烯、乳胶,等)珠、以及用于结合上述标记物修饰的亲和素(例如,链霉亲和素)的生物素。
在某些优选的实施方案中,所述第一试剂通过免疫学检测来测定所述样品中OAS2或USP18的蛋白水平。在某些优选的实施方案中,所述免疫学检测选自ELISA检测、Elispot检测、Western印迹或表面等离子共振法。在某些实施方式中,所述第一试剂包括抗OAS2或USP18蛋白的抗体或其抗原结合片段。
在某些优选的实施方案中,本发明的试剂盒包含能够检测OAS2表达水平的第一试剂,任选地还包括IFNα和/或刺激物。
在某些优选的实施方案中,本发明的试剂盒包含能够检测USP18表达水平的第一试剂和IFNα,任选地还包括刺激物。
在某些优选的实施方案中,本发明的试剂盒还包含用于预处理含有PBMC的样品的第二试剂,其中所述第二试剂包括选自下列的一种或多种试剂:用于稀释样品的稀释剂(例如,磷酸盐缓冲液或生理盐水);用于阻止血液凝固的抗凝剂(例如,肝素);用于维持
PBMC活性的试剂(例如,培养基或培养液);和,用于分离PBMC的试剂(例如,淋巴细胞分离液,例如聚蔗糖-泛影葡胺溶液)。在某些优选的实施方案中,所述第二试剂包括用于维持PBMC活性的试剂(例如,培养基或培养液),和/或用于分离PBMC的试剂(例如,淋巴细胞分离液,例如聚蔗糖-泛影葡胺溶液)。
在某些优选的实施方案中,本发明的试剂盒还包含采血装置(例如无热原真空采血管)和/或PBMC分离装置(例如PBMC细胞分离管)。
在某些优选的实施方案中,本发明的试剂盒还包含能够检测内参基因表达水平的试剂,所述内参基因在各组织和细胞中的表达相对恒定,并在处理因素作用的条件下不会发生表达改变,例如管家基因;通常,所述管家基因编码对维持细胞基本生命活动所必需的蛋白质,其包括但不限于β-actin、GAPDH、18S rRNA、β2-MG、UBC或α-tubulin。在某些优选的实施方案中,所述能够检测内参基因表达水平的试剂为能够检测所述内参基因的mRNA水平的试剂。在某些优选的实施方案中,所述能够检测内参基因表达水平的试剂为能够检测所述内参基因的蛋白水平的试剂。
在另一个方面,本发明涉及能够检测标志物表达水平的试剂在制备试剂盒中的用途,所述试剂盒用于预测IFNα对患有慢性乙肝的受试者的治疗效果或所述受试者对IFNα治疗的应答;其中,所述标志物选自OAS2或USP18。
在某些优选的实施方案中,所述试剂盒还包括IFNα和/或刺激物,所述刺激物选自HBV抗原、HBV抗原的抗原性片段或其任何组合。
在某些优选的实施方案中,所述HBV抗原为HBcAg。在某些优选的实施方案中,所述HBcAg具有如SEQ ID NO:38所示的氨基酸序列。在某些优选的实施方案中,所述抗原性片段具有选自下列的氨基酸序列:SEQ ID NO:3-37。
在一个特别优选的实施方案中,所述刺激物包含分别具有如SEQ ID NO:3-37所示的氨基酸序列的抗原性片段。
在某些优选的实施方案中,所述能够检测标志物表达水平的试剂为能够检测所述标志物的mRNA水平的试剂。这类试剂是本领域公知的,包括但不限于特异性结合目标序列的核酸探针、扩增目标序列的引物、非特异性荧光染料(例如,SYBR Green I)或其组合。在某些实施方式中,所述核酸探针可以为单标记的核酸探针,例如放射性核素(如32P、3H、35S等)标记探针、生物素标记探针、辣根过氧化物酶标记探针、地高辛标记探
针或荧光基团(如FITC、FAM、TET、HEX、TAMRA、Cy3、Cy5等)标记探针;所述核酸探针也可以为双标记的核酸探针,例如Taqman探针、分子信标、置换探针、蝎子引物探针、QUAL探针、FRET探针等。在某些实施方式中,所述能够检测标志物表达水平的试剂包括Taqman探针。在某些实施方式中,所述OAS2的cDNA具有如SEQ ID NO:1所示的核苷酸序列。在某些实施方式中,所述USP18的cDNA具有如SEQ ID NO:2所示的核苷酸序列。
可以采用本领域已知的mRNA检测方法测定所述标志物的mRNA水平,例如荧光定量PCR,Northern印迹法,原位杂交法,或包括mRNA扩增(例如反转录PCR)和所述mRNA扩增产物的定量(例如电泳和染色)的方法。在某些优选的实施方案中,所述能够检测标志物表达水平的试剂通过PCR定量测定所述标志物的mRNA水平。在一个具体的实施方式中,所述能够检测标志物表达水平的试剂通过荧光定量PCR测定所述标志物的mRNA水平。
在某些优选的实施方案中,所述能够检测标志物表达水平的试剂为能够检测所述标志物的蛋白水平的试剂。这类试剂是本领域熟知的,包括但不限于能够和OAS2或USP18蛋白特异性结合的抗体、靶向多肽或核酸适体。在某些实施方式中,这类试剂带有可检测的标记,例如酶(如辣根过氧化物酶、碱性磷酸酶等)、放射性核素(如3H、125I、35S、14C、32P等)、荧光染料(如FITC、TRITC、PE、Texas Red、量子点、Cy7、Alexa 750等)、吖啶酯类化合物、磁珠(例如,)、胶体金或有色玻璃或塑料(例如,聚苯乙烯、聚丙烯、乳胶,等)珠、以及用于结合上述标记物修饰的亲和素(例如,链霉亲和素)的生物素。
在某些优选的实施方案中,所述能够检测标志物表达水平的试剂通过免疫学检测来测定所述样品中OAS2或USP18的蛋白水平。在某些优选的实施方案中,所述免疫学检测选自ELISA检测、Elispot检测、Western印迹或表面等离子共振法。在某些实施方式中,所述能够检测标志物表达水平的试剂包括抗OAS2或USP18蛋白的抗体或其抗原结合片段。
在某些优选的实施方案中,所述试剂盒还包含能够检测内参基因表达水平的试剂,所述内参基因在各组织和细胞中的表达相对恒定,并在处理因素作用的条件下不会发生表达改变,例如管家基因;通常,所述管家基因编码对维持细胞基本生命活动所必需的蛋白质,其包括但不限于β-actin、GAPDH、18S rRNA、β2-MG、UBC或α-tubulin。在某些优选的实施方案中,所述能够检测内参基因表达水平的试剂为能够检测所述内参基
因的mRNA水平的试剂。在某些优选的实施方案中,所述能够检测内参基因表达水平的试剂为能够检测所述内参基因的蛋白水平的试剂。
在某些优选的实施方案中,所述试剂盒还包含一种或多种选自1)-6)的试剂或装置:
1)用于稀释样品的稀释剂,例如磷酸盐缓冲液或生理盐水;
2)用于阻止血液凝固的抗凝剂,例如肝素;
3)用于维持PBMC活性的试剂,例如培养基或培养液;
4)用于分离PBMC的试剂,例如淋巴细胞分离液,例如聚蔗糖-泛影葡胺溶液;
5)采血装置,例如无热原真空采血管;和
6)PBMC分离装置,例如PBMC细胞分离管。
在某些优选的实施方案中,所述试剂盒包括用于维持PBMC活性的试剂(例如,培养基或培养液),和/或用于分离PBMC的试剂(例如,淋巴细胞分离液,例如聚蔗糖-泛影葡胺溶液)。
在某些优选的实施方案中,所述试剂盒通过包括下述步骤的方法来预测IFNα对患有慢性乙肝的受试者的治疗效果或所述受试者对IFNα治疗的应答:
(1)使用能够检测标志物表达水平的试剂测定来自所述受试者的样品中所述标志物的表达水平,其中所述标志物为OAS2;和
(2)将该表达水平与参考值进行比较,或对该表达水平进行统计学分析以获得统计分析值,并将该统计分析值与参考值进行比较,并判断所述受试者是否会对IFNα治疗产生应答;
其中,所述样品包含外周血单个核细胞(PBMC),例如全血(例如抗凝全血)、外周血单个核细胞(PBMC)、或外周血白膜层。
在某些优选的实施方案中,当所述表达水平大于所述参考值时,或当所述表达水平的统计分析值大于所述参考值时,表明所述受试者会对IFNα治疗产生应答,适宜接受IFNα治疗;当所述表达水平不大于所述参考值时,或当所述表达水平的统计分析值不大于所述参考值时,表明所述受试者不会对IFNα治疗产生应答,不适宜接受IFNα治疗。
在某些优选的实施方案中,在步骤(2)中,使用Logistic回归模型对所述表达水平进行统计分析。
在某些优选的实施方案中,为了消除样品与样品之间的待测标志物起始值的差异,以使样品之间具有可比性,通常可以对不同样品中待测标志物的表达水平值进行归一化
处理,例如将样品中待测标志物的表达水平值与内参基因的表达水平值进行比较。因此,在步骤(1)中,所述标志物的表达水平为归一化表达水平。所述归一化表达水平可通过以下示例性方法获得:使用能够检测内参基因表达水平的试剂测定来自所述受试者的样品中所述内参基因的表达水平,并将所述标志物的表达水平与所述内参基因的表达水平进行比较,以获得所述标志物的归一化表达水平。在一个具体的实施方式中,在步骤(1)中,所述标志物的表达水平为归一化mRNA水平,其可通过以下示例性方法获得:使用比较Ct法将待测样品中所述标志物的mRNA水平与所述内参基因的mRNA水平进行比较,获得2-ΔCt值(ΔCt=Ct待测标志物-Ct内参基因),该值即为所述标志物的归一化mRNA水平,也即所述标志物的表达量相比于内参基因的表达量的变化倍数。
在某些优选的实施方案中,在步骤(1)之前,所述方法还包括下列步骤中的一项或多项:(a)使用采血装置从所述受试者获得样品;(b)使用抗凝剂处理采血装置或来自所述受试者的样品;(c)使用用于维持PBMC活性的试剂处理来自所述受试者的样品;(d)使用稀释剂稀释来自所述受试者的样品;和(e)使用用于分离PBMC的试剂或PBMC分离装置从来自所述受试者的样品中分离获得PBMC。
在某些优选的实施方案中,所述试剂盒通过包括下述步骤的方法来预测IFNα对患有慢性乙肝的受试者的治疗效果或所述受试者对IFNα治疗的应答:
(1)使用IFNα刺激来自所述受试者的至少一份样品作为待测样品,并将未经IFNα刺激的样品作为对照样品;或,使用IFNα和刺激物共同刺激来自所述受试者的至少一份样品作为待测样品,并将经所述刺激物刺激但未经IFNα刺激的样品作为对照样品;其中,所述刺激物选自HBV抗原、HBV抗原的抗原性片段或其任何组合;
(2)使用能够检测标志物表达水平的试剂测定步骤(1)中各个样品中所述标志物的表达水平,并获得所述待测样品中该标志物的表达水平相比于所述对照样品中该标志物的表达水平的变化倍数,将该变化倍数作为所述待测样品中所述标志物的相对表达水平,其中所述标志物选自OAS2或USP18;和
(3)将该相对表达水平与参考值进行比较,或对该相对表达水平进行统计学分析以获得统计分析值,并将该统计分析值与参考值进行比较,并判断所述受试者是否会对IFNα治疗产生应答;
其中,所述样品包含外周血单个核细胞(PBMC),例如全血(例如抗凝全血)、外周血单个核细胞(PBMC)、或外周血白膜层。
在某些优选的实施方案中,当所述相对表达水平小于所述参考值时,或当所述相对表达水平的统计分析值小于所述参考值时,表明所述受试者会对IFNα治疗产生应答,适宜接受IFNα治疗;当所述相对表达水平不小于所述参考值时,或当所述相对表达水平的统计分析值不小于所述参考值时,表明所述受试者不会对IFNα治疗产生应答,不适宜接受IFNα治疗。
在某些优选的实施方案中,在步骤(2)中,通过比较Ct法获得所述待测样品中该标志物的mRNA水平相比于所述对照样品中该标志物的mRNA水平的变化倍数,即所述待测样品中所述标志物的mRNA相对表达水平。
在某些优选的实施方案中,在步骤(3)中,使用Logistic回归模型对所述相对表达水平进行统计分析。
在某些优选的实施方案中,为了消除样品与样品之间的待测标志物起始值的差异,以使样品之间具有可比性,通常可以对不同样品中待测标志物的表达水平值进行归一化处理,例如将样品中待测标志物的表达水平值与内参基因的表达水平值进行比较。因此,在步骤(2)中,所述标志物的表达水平为归一化表达水平。所述归一化表达水平可通过以下示例性方法获得:使用能够检测内参基因表达水平的试剂测定来自所述受试者的样品中所述内参基因的表达水平,并将所述标志物的表达水平与所述内参基因的表达水平进行比较,以获得所述标志物的归一化表达水平。在一个具体的实施方式中,在步骤(2)中,所述标志物的表达水平为归一化mRNA水平,其可通过以下示例性方法获得:使用比较Ct法将待测样品中所述标志物的mRNA水平与所述内参基因的mRNA水平进行比较,获得所述标志物的归一化mRNA水平。进一步,通过比较Ct法获得所述待测样品中该标志物的均一化mRNA水平相比于所述对照样品中该标志物的均一化mRNA水平的变化倍数,即所述待测样品中所述标志物的mRNA相对表达水平。
在某些优选的实施方案中,在步骤(1)之前,所述方法还包括下列步骤中的一项或多项:(a)使用采血装置从所述受试者获得样品;(b)使用抗凝剂处理采血装置或来自所述受试者的样品;(c)使用用于维持PBMC活性的试剂处理来自所述受试者的样品;(d)使用稀释剂稀释来自所述受试者的样品;和(e)使用用于分离PBMC的试剂或PBMC分离装置从来自所述受试者的样品中分离获得PBMC。
在另一个方面,本发明提供了用于预测IFNα对患有慢性乙肝的受试者的治疗效果
或所述受试者对IFNα治疗的应答的方法,或获得诊断分析的结果的方法,其中所述诊断分析用于预测IFNα对患有慢性乙肝的受试者的治疗效果或所述受试者对IFNα治疗的应答,其包括下述步骤:
(1)提供来自所述受试者的样品;
(2)测定所述样品中标志物的表达水平,其中所述标志物为OAS2;和
(3)将该表达水平与参考值进行比较,或对该表达水平进行统计学分析以获得统计分析值,并将该统计分析值与参考值进行比较,并判断所述受试者是否会对IFNα治疗产生应答;
其中,所述样品包含外周血单个核细胞(PBMC),例如全血(例如抗凝全血)、外周血单个核细胞(PBMC)、或外周血白膜层。
在某些优选的实施方式中,在步骤(3)之后,所述方法还包括给所述受试者施用IFNα以治疗慢性乙肝的步骤,其中所述受试者被判断为会对IFNα治疗产生应答。
在某些优选的实施方案中,当所述表达水平大于所述参考值时,或当所述表达水平的统计分析值大于所述参考值时,表明所述受试者会对IFNα治疗产生应答,适宜接受IFNα治疗;当所述表达水平不大于所述参考值时,或当所述表达水平的统计分析值不大于所述参考值时,表明所述受试者不会对IFNα治疗产生应答,不适宜接受IFNα治疗。
在某些优选的实施方案中,在步骤(2)中,测定所述标志物的mRNA水平或蛋白水平。
在某些优选的实施方案中,在步骤(2)中,测定所述标志物的mRNA水平。可以采用本领域已知的mRNA检测方法测定所述标志物的mRNA水平,例如荧光定量PCR,Northern印迹法,原位杂交法,或包括mRNA扩增(例如反转录PCR)和所述mRNA扩增产物的定量(例如电泳和染色)的方法。在某些优选的实施方案中,通过PCR定量测定所述标志物的mRNA水平。在一个具体的实施方式中,通过荧光定量PCR测定所述标志物的mRNA水平。在某些实施方式中,所述OAS2的cDNA具有如SEQ ID NO:1所示的核苷酸序列。在某些实施方式中,所述USP18的cDNA具有如SEQ ID NO:2所示的核苷酸序列。
在某些优选的实施方案中,在步骤(2)中,测定所述标志物的蛋白水平。在某些优选的实施方案中,在步骤(2)中,通过免疫学检测来测定所述标志物的蛋白水平。进一步,在某些优选的实施方案中,所述免疫学检测选自ELISA检测、Elispot检测、Western印迹或表面等离子共振法。在某些实施方式中,在步骤(2)中,使用抗OAS2的抗体或
其抗原结合片段来检测OAS2的蛋白水平。
在某些优选的实施方案中,在步骤(3)中,使用Logistic回归模型对所述表达水平进行统计分析。
在某些优选的实施方案中,为了消除样品与样品之间的待测标志物起始值的差异,以使样品之间具有可比性,通常可以对不同样品中待测标志物的表达水平值进行归一化处理,例如将样品中待测标志物的表达水平值与内参基因的表达水平值进行比较。因此,在步骤(2)中,所述标志物的表达水平为归一化表达水平。所述归一化表达水平可通过以下示例性方法获得:使用能够检测内参基因表达水平的试剂测定来自所述受试者的样品中所述内参基因的表达水平,并将所述标志物的表达水平与所述内参基因的表达水平进行比较,以获得所述标志物的归一化表达水平。在一个具体的实施方式中,在步骤(2)中,所述标志物的表达水平为归一化mRNA水平,其可通过以下示例性方法获得:使用比较Ct法将待测样品中所述标志物的mRNA水平与所述内参基因的mRNA水平进行比较,获得2-ΔCt值(ΔCt=Ct待测标志物-Ct内参基因),该值即为所述标志物的归一化mRNA水平,也即所述标志物的表达量相比于内参基因的表达量的变化倍数。
在某些优选的实施方案中,在步骤(1)之前,还包括下列步骤中的一项或多项:(a)从所述受试者获得样品;(b)向来自所述受试者的样品中加入抗凝剂,例如肝素;(c)从来自所述受试者的样品中获取PBMC或含有PBMC的血液成分(例如,外周血白膜层);(d)向来自所述受试者的样品中加入培养液或培养基;和,(e)稀释来自所述受试者的样品。
在另一个方面,本发明提供了用于预测IFNα对患有慢性乙肝的受试者的治疗效果或所述受试者对IFNα治疗的应答的方法,或获得诊断分析的结果的方法,其中所述诊断分析用于预测IFNα对患有慢性乙肝的受试者的治疗效果或所述受试者对IFNα治疗的应答,其包括下述步骤:
(1)提供来自所述受试者的至少两份样品;
(2)使用IFNα刺激至少一份样品作为待测样品,并将未经IFNα刺激的样品作为对照样品;或,使用IFNα和刺激物共同刺激来自所述受试者的至少一份样品作为待测样品,并将经所述刺激物刺激但未经IFNα刺激的样品作为对照样品;其中,所述刺激物选自HBV抗原、HBV抗原的抗原性片段或其任何组合;
(3)测定步骤(2)中各个样品中标志物的表达水平,并获得所述待测样品中该标志物的表达水平相比于所述对照样品中该标志物的表达水平的变化倍数,将该变化倍数作为所述待测样品中所述标志物的相对表达水平,其中所述标志物选自OAS2或USP18;和
(4)将该相对表达水平与参考值进行比较,或对该相对表达水平进行统计学分析以获得统计分析值,并将该统计分析值与参考值进行比较,并判断所述受试者是否会对IFNα治疗产生应答;
其中,所述样品包含外周血单个核细胞(PBMC),例如全血(例如抗凝全血)、外周血单个核细胞(PBMC)、或外周血白膜层。
在某些优选的实施方式中,在步骤(4)之后,所述方法还包括给所述受试者施用IFNα以治疗慢性乙肝的步骤,其中所述受试者被判断为会对IFNα治疗产生应答。
在某些优选的实施方案中,当所述相对表达水平小于所述参考值时,或当所述相对表达水平的统计分析值小于所述参考值时,表明所述受试者会对IFNα治疗产生应答,适宜接受IFNα治疗;当所述相对表达水平不小于所述参考值时,或当所述相对表达水平的统计分析值不小于所述参考值时,表明所述受试者不会对IFNα治疗产生应答,不适宜接受IFNα治疗。
在某些优选的实施方案中,在步骤(2)中,所述HBV抗原为HBVcAg。在某些优选的实施方案中,所述HBVcAg具有如SEQ ID NO:38所示的氨基酸序列。在某些优选的实施方案中,所述抗原性片段具有选自下列的氨基酸序列:SEQ ID NO:3-37。在一个特别优选的实施方案中,所述刺激物包含分别具有如SEQ ID NO:3-37所示的氨基酸序列的抗原性片段。
在某些优选的实施方案中,在步骤(3)中,测定所述标志物的mRNA水平。可以采用本领域已知的mRNA检测方法测定所述标志物的mRNA水平,例如荧光定量PCR,Northern印迹法,原位杂交法,或包括mRNA扩增(例如反转录PCR)和所述mRNA扩增产物的定量(例如电泳和染色)的方法。在某些优选的实施方案中,通过PCR定量测定所述标志物的mRNA水平。在一个具体的实施方式中,通过荧光定量PCR测定所述标志物的mRNA水平。在某些实施方式中,所述OAS2的cDNA具有如SEQ ID NO:1所示的核苷酸序列。在某些实施方式中,所述USP18的cDNA具有如SEQ ID NO:2所示的核苷酸序列。
在某些优选的实施方案中,在步骤(3)中,测定所述标志物的蛋白水平。在某些优
选的实施方案中,在步骤(3)中,通过免疫学检测来测定所述标志物的蛋白水平。进一步,在某些优选的实施方案中,所述免疫学检测选自ELISA检测、Elispot检测、Western印迹或表面等离子共振法。在某些实施方式中,在步骤(3)中,使用抗OAS2或USP18蛋白的抗体或其抗原结合片段来检测所述标志物的蛋白水平。
在某些优选的实施方案中,在步骤(3)中,通过比较Ct法获得所述待测样品中该标志物的mRNA水平相比于所述对照样品中该标志物的mRNA水平的变化倍数,即所述待测样品中所述标志物的mRNA相对表达水平。
在某些优选的实施方案中,在步骤(4)中,使用Logistic回归模型对所述相对表达水平进行统计分析。
在某些优选的实施方案中,为了消除样品与样品之间的待测标志物起始值的差异,以使样品之间具有可比性,通常可以对不同样品中待测标志物的表达水平值进行归一化处理,例如将样品中待测标志物的表达水平值与内参基因的表达水平值进行比较。因此,在步骤(3)中,所述标志物的表达水平为归一化表达水平。所述归一化表达水平可通过以下示例性方法获得:使用能够检测内参基因表达水平的试剂测定来自所述受试者的样品中所述内参基因的表达水平,并将所述标志物的表达水平与所述内参基因的表达水平进行比较,以获得所述标志物的归一化表达水平。在一个具体的实施方式中,在步骤(3)中,所述标志物的表达水平为归一化mRNA水平,其可通过以下示例性方法获得:使用比较Ct法将待测样品中所述标志物的mRNA水平与所述内参基因的mRNA水平进行比较,获得所述标志物的归一化mRNA水平。进一步,通过比较Ct法获得所述待测样品中该标志物的均一化mRNA水平相比于所述对照样品中该标志物的均一化mRNA水平的变化倍数,即所述待测样品中所述标志物的mRNA相对表达水平。
在某些优选的实施方案中,在步骤(1)之前,还包括下列步骤中的一项或多项:(a)从所述受试者获得样品;(b)向来自所述受试者的样品中加入抗凝剂,例如肝素;(c)从来自所述受试者的样品中获取PBMC或含有PBMC的血液成分(例如,外周血白膜层);(d)向来自所述受试者的样品中加入培养液或培养基;和,(e)稀释来自所述受试者的样品。
本发明还包含以下示例性实施方案:
1.用于体外诱导干扰素诱导基因(Interferon-stimulated genes,ISGs)的刺激
物,其包含HBV核心抗原的全长或部分片段的组合和/或IFNα。
2.用于预测慢性乙肝患者α干扰素治疗效果的标志物,其中所述标志物为干扰素诱导基因(Interferon-stimulated genes,ISGs)。
3.项目2的标志物,其中所述干扰素诱导基因选自OAS2和USP18。
4.筛选用于预测慢性乙肝患者α干扰素治疗效果的标志物的方法,所述方法包括下述步骤:
1)收集慢性乙肝患者用药前全血标本,分离PBMC;
2)用HBV核心抗原多肽段或IFNα或HBV核心抗原多肽段与IFNα的组合诱导PBMC样品;
3)收集诱导后的PBMC样品,提取RNA,反转录后检测ISGs mRNA水平;
4)收集标本临床IFNα治疗应答结果,筛选ISGs;
其中,比较IFNα治疗应答组和无应答组相同诱导条件下不同ISGs mRNA水平,IFNα治疗应答组和无应答组ISGs mRNA水平高低的不同可以作为治疗效果的依据。
5.项目4的方法,其中所述HBV核心抗原多肽段是HBV核心抗原的第1-183位氨基酸的多肽段。
6.用于预测慢性乙肝患者α干扰素治疗效果的试剂盒,其包含HBV核心抗原多肽段和IFNα诱导物,和检测标志物OAS2和USP18水平的试剂。
7.项目6的试剂盒,其中所述HBV核心抗原多肽段是HBV核心抗原的第1-183位氨基酸的多肽段。
8.检测标志物OAS2和/或USP18水平的试剂在制备用于预测慢性乙肝患者α干扰素治疗效果的诊断剂中的用途。
9.检测标志物OAS2和/或USP18水平的试剂在制备用于预测慢性乙肝患者α干扰素治疗效果的试剂盒中的用途。
10.标志物OAS2和/或USP18用于预测慢性乙肝患者α干扰素治疗效果的用途。
11.项目8-10任一项中的用途,其中所述标志物OAS2和/或USP18的检测包括定量检测标志物OAS2和/或USP18的mRNA水平。
12.项目8-10任一项中的用途,其中通过荧光PCR定量检测标志物OAS2和/或USP18的mRNA水平。
13.项目8-10任一项中的用途,其中用于检测标志物OAS2和/或USP18的mRNA水
平的引物和探针的序列为:
发明的有益效果
本发明通过大量实验和反复摸索发现了OAS2和/或USP18的表达水平在对IFNα治疗应答和对IFNα治疗无应答的CHB患者这两个群体中,存在显著差异,由此建立了一种简单、方便、快速且具有高准确性和特异性的预测IFNα对患有慢性乙肝的受试者的治疗效果或所述受试者对IFNα治疗的应答的预测方法。
与现有技术相比,本发明的技术方案具有以下有益效果:
(1)首次发现慢性乙肝患者PBMC未经或经体外刺激后的OAS2表达水平能够用于预测CHB患者IFNα治疗的结果;
(2)首次发现慢性乙肝患者PBMC经体外刺激后USP18的表达水平能够用于预测CHB患者IFNα治疗的结果;
(3)所涉及的操作简单、方便:例如,通过采集全血、分离PBMC,将其分装至细胞培养板并添加IFNα后,然后置于恒温培养箱中培养;培养一定时间后进行mRNA或蛋白水平的检测;
(4)对实验条件、人员的技术能力、设备和环境的要求不高;
(5)对IFNα治疗应答结果预测准确率高;
(6)成本不高,易于推广,使用范围广。
下面将结合附图和实施例对本发明的实施方案进行详细描述,但是本领域技术人员将理解,下列附图和实施例仅用于说明本发明,而不是对本发明的范围的限定。根据附图和优选实施方案的下列详细描述,本发明的各种目的和有利方面对于本领域技术人员来说将变得显然。
图1显示了在不同处理条件下,CHB患者的PBMC的OAS2mRNA水平在对IFNα治疗应答组(Rs)和无应答组(NRs)之间存在显著差异。其中,图1a显示PBMC不经体外诱导,应答组OAS2的mRNA水平显著高于无应答组。图1b显示PBMC经IFNα体外诱导后,应答组OAS2的mRNA相对表达水平显著低于无应答组。(注:*表示p<0.05,****表示p<0.0005)
图2显示了在不同处理条件下,CHB患者的PBMC的USP18mRNA水平在对IFNα治疗应答组(Rs)和无应答组(NRs)之间存在显著差异。其中,图2a显示PBMC经IFNα体外诱导后,应答组USP18的mRNA相对表达水平显著低于无应答组;;图2b显示PBMC经IFNα和HBcAg多肽库共同诱导后,应答组USP18的mRNA相对表达水平显著低于无应答组。(注:料*表示p<0.001,料**表示p<0.0005)
图3显示了OAS2N检测法和OAS2(IFNa+N-N)检测法的ROC曲线。结果显示,OAS2N检测法和OAS2(IFNa+N-N)检测法对于预测CHB患者对IFNα治疗的应答均具有良好的灵敏度和特异性。
图4显示了OAS2(IFNa+N-N)检测法、USP18(IFNa+N-N)检测法及两者联合检测的ROC曲线。结果显示,OAS2(IFNa+N-N)检测法、USP18(IFNa+N-N)检测法及两者的联合检测对于预测CHB患者对IFNα治疗的应答具有很好的灵敏度和特异性。
序列信息
本发明涉及的序列信息提供于下面的表1中。
表1:序列信息
现参照下列意在举例说明本发明(而非限定本发明)的实施例来描述本发明。除非特别指明,否则基本上按照本领域内熟知的以及在各种参考文献中描述的常规方法进行
实施例中描述的实验和方法(例如,分子生物学实验方法和免疫检测法)。参见,例如,Sambrook等人,Molecular Cloning:A Laboratory Manual,第2版,Cold Spring Harbor Laboratory Press,Cold Spring Harbor,N.Y.(1989);和Ausubel等人,Current Protocols in Molecular Biology,Greene Publishing Associates(1992),其全部通过引用合并入本文。所用试剂或仪器未注明生产厂商者,均为可以通过市购获得的常规产品。本领域技术人员知晓,实施例以举例方式描述本发明,且不意欲限制本发明所要求保护的范围。本文中提及的全部公开案和其他参考资料以其全文通过引用合并入本文。
实施例1.外周血PBMC分离和体外诱导培养
1.1外周血PBMC分离
(1)将全血在4000rpm条件下离心10min。
(2)将血清吸出,加等量的PBS稀释余下的全血。
(3)准备15mL的离心管,加入与全血等量的淋巴细胞分离液(Ficoll-page Tm PLUS,GE Helathcare Life Sciences)。
(4)将稀释后的全血沿管壁缓慢加在淋巴细胞分离液之上,保持分层状态。
(5)将上述液体在1000g条件下离心22min,并将升速和降速的速率调到最小。
(6)小心吸取白色云雾状淋巴细胞层,至另一离心管中,加入PBS稀释后离心,600g×7min,弃上清。再加入PBS稀释,离心400g×7min,弃上清。用1ml的培养液重悬细胞。
(7)在1.5ml离心管中加入10μl已制备样品细胞悬液和40μl的0.4%台盼蓝,制备1:5细胞稀释液。取10μl台盼蓝/细胞混合悬液加入血细胞计数板并进行活细胞计数,其中每毫升细胞数量=细胞计数×稀释倍数×104。
1.2IFNα和/或刺激物对PBMC的体外诱导培养
将上述PBMC细胞接种于24孔板内,每孔加入5×105个细胞,并添加1ml含10%胎牛血清的RPM1640培养基。每份标本共设置四个孔,其中第一孔不添加诱导物质(所述诱导物质包括IFNα和HBcAg多肽库)标记为N,第二孔添加IFNα(安达芬,安徽安科生物工程(集团)股份有限公司)(1×104IU/mL),第三孔添加含有35条HBcAg抗原性片段(SEQ ID NO:3-37)的多肽库(由上海生物工程股份有限公司合成)(1μg/条/mL,共35条),第四孔添加上述IFNα和HBcAg多肽库。将培养板置于37℃二氧化碳培养箱
中诱导培养24h后,收集细胞。
实施例2.荧光定量PCR法检测基因的mRNA水平
本发明对包括USP18、OAS2、LGP1、LAP3、GIP3、CEB1、ATF5、GIP2、OAS3、RPLP2、STXBP5、Viperin、RPS28、PI3KAP1、MX1、DUSP1、ETEF1等17种基因和内参β-actin建立了的荧光定量PCR检测体系。
2.1PBMC中RNA的提取
(1)离心收集实施例1中所获得PBMC细胞,弃去上清;每孔添加250μl RPMI1640培养基和750μl Tripure,并混合均匀;每1ml细胞中添加200μl氯仿,上下颠倒混匀,4℃静置10min。
(2)12,000rpm,4℃,离心15min。
(3)经上述处理后,每管内样品共分为三层,吸取最上面一层(注意不要吸到蛋白层)转移到一个新的用DEPC处理过的EP管中,加等体积异丙醇,上下颠倒混匀,-20℃静置15min。
(4)12,000rpm,4℃,离心10min,弃上清。
(5)加入1mL 75%乙醇洗两遍,12,000rpm,4℃,离心5min。
(6)弃去乙醇,空气风干5-10min,用55-60℃预热好的DEPC水35μL溶解RNA,提取好的RNA冻存于-80℃冰箱保存。
2.2RNA反转录
采用分光光度计测定RNA样品的浓度和纯度。,取2μL RNA样在2μL RNase-free water校准仪器中测定A260与A280吸光值,检测A260与A280比值从而评价RNA样品的纯度,经检测,RNA纯度:A260/280≥1.80。
反转录体系:
通过上述反转录体系,在42℃反转录30min。获得的cDNA即为荧光定量PCR检测体系中的模板,保存于-20℃。
2.3荧光定量PCR体系建立
(1)RT-PCR体系:
扩增反应过程:
95℃3min,然后95℃10sec,55℃45sec,共39个循环。
应用primer软件设计用于RT-PCR的检测引物及探针,并由英骏生物技术有限公司合成,具体序列见下表2:
表2.用于荧光定量PCR反应的引物和探针
(2)引物预实验
对用primer软件设计的18套引物(上文表2)进行预实验,随机选取两份来自CHB患者的模板(分别标记为CHB 1、CHB 2),通过荧光定量PCR检测待测基因及内参引物的Ct值。鉴定结果如表3所示,17套待测基因的引物和内参β-actin引物Ct值均在15-35之间,表明其均能进行有效扩增;同时Neg组(不含模板)未检测到有效扩增,进一步表明上文表2中所列举的18套引物可用于相应的待测基因的荧光定量PCR检测。
表3.荧光定量PCR检测体系中引物的预实验
实施例3.OAS2、USP18在对IFNα治疗应答和无应答的CHB患者中的表达差异
本发明共临床追踪了150例慢性乙肝标本,其中接受IFNα治疗的且有24周治疗结果的共66例;其中对IFNα治疗无应答共47例,对IFNα治疗应答共19例。表4显示
了本实施例所入选研究样本的特征数据,结果显示,应答组(Rs)和无应答组(NRs)在年龄、性别上没有显著差异,治疗前ALT、AST和HBsAg、HBV DNA、HBeAg结果也没有显著差异。
表4.入选研究标本特征
对还未接受IFNα治疗的慢性乙肝患者采集外周血,按照上述实施例1的方法提取PBMC并对PBMC进行处理和实施例2的方法提取RNA,进行荧光定量PCR以检测计算17个待测基因(USP18、OAS2、LGP1、LAP3、GIP3、CEB1、ATF5、GIP2、OAS3、RPLP2、STXBP5、Viperin、RPS28、PI3KAP1、MX1、DUSP1、ETEF1)的mRNA水平,所述mRNA水平通过相对定量法(比较Ct法)计算得到,其计算方式为:将测得的每个目的基因的Ct值减去各自内参β-actin的Ct值,得到每个目的基因的ΔCt值(ΔCt=Ct目的基因-Ct内参基因),计算2-ΔCt值,其所表示的是待测样本中该基因的表达量相对于内参基因的表达量的变化倍数(即,该基因相对于内参基因的归一化mRNA水平);然后将待测样本中该基因的ΔCt值与对照样本中该基因的ΔCt值相减,得到ΔΔCt值(ΔΔCt=ΔCt待测样本-ΔCt对照样本),最后计算2-ΔΔCt值,其所表示的是待测样本该基因的表达量相对于对照样本该基因的表达量的变化倍数(即,待测样本中该基因相对于对照样本中该基因的mRNA相对表达水平)。
通过比较应答组和无应答组中上述各个基因的mRNA水平的差异,发现其中OAS2和USP18的mRNA水平在应答组和无应答组之间存在明显差异。结果如图1和图2所示,其中OAS2N组的PBMC细胞未经任何体外诱导,该组所表示的是待测样本中OAS2相对于内参基因的归一化mRNA水平;OAS2(IFNa+N-N)组、USP18(IFNa+N-N)组和USP18(IFNa+HBcAg-HBcAg)组的PBMC体外诱导条件如表5所示,其体外诱导培养的具体步骤参照实施例1.2进行,其中,OAS2(IFNa+N-N)组所表示的是待测样本中OAS2相对于对照样本中OAS2的mRNA相对表达水平;USP18(IFNa+N-N)组和USP18(IFNa+HBcAg-HBcAg)组所分别表示的是待测样本中USP18相对于各自对照样本中USP18的mRNA相对表达水平。
表5.不同组别内待测样本及对照样本的体外诱导条件
图1a显示了OAS2N组的检测结果,结果显示,未经任何体外诱导的CHB患者接受IFNα治疗之前的PBMC样品中,应答组患者的OAS2归一化mRNA水平显著高于无应答组患者。图1b显示了OAS2(IFNa+N-N)组的检测结果,结果显示,经IFNα体外诱导培养的CHB患者接受IFNα治疗之前的PBMC样品中,应答组患者的OAS2的mRNA相对表达水平显著低于无应答组患者。
图2a显示了USP18(IFNa+N-N)组的检测结果,结果显示,经IFNα体外诱导培养的CHB患者接受IFNα治疗之前的PBMC样品中,应答组患者的USP18的mRNA相对表达水平显著低于无应答组患者。图2b显示了USP18(IFNa+HBcAg-HBcAg)组的检测结果,结果显示,经IFNα和HBcAg多肽库(含有SEQ ID NO:3-37所示的HBcAg抗原性片段)体外诱导培养的CHB患者接受IFNα治疗之前的PBMC样品中,应答组患者的USP18的mRNA相对表达水平显著低于无应答组患者。
上述结果表明,对IFNα治疗应答患者的治疗前PBMC样品中OAS2水平相比于对IFNα治疗无应答患者显著升高;同时,对IFNα治疗应答患者的治疗前PBMC样本经IFNα和/或HBcAg体外诱导后其中OAS2和/或UPS18水平相比于对IFNα治疗无应答患者显著降低。因此OAS2和/或UPS18可以用于预测CHB患者对IFNα治疗应答。
实施例4.OAS2N检测法、OAS2(IFNa+N-N)检测法的ROC曲线分析
根据实施例3中OAS2N组和OAS2(IFNa+N-N)组的实验条件,分别建立了相应的OAS2N检测法和OAS2(IFNa+N-N)检测法,表6记载了上述两种方法的主要步骤。本实施例采用了如上所述的两种方法分别对实施例3中所记载的CHB患者样本进行了检测,分别获得样本的两个诊断指标:OAS2归一化mRNA水平和OAS2的mRNA相对表达水平;然后使用SPSS 17.0软件以OAS2归一化mRNA水平和OAS2的mRNA相对表达水平为诊断指标,绘制ROC曲线以检验单独使用OAS2N检测法或OAS2(IFNa+N-N)检测法在预测IFNα对CHB患
者的治疗效果或CHB患者对IFNα治疗的应答方面的性能。ROC曲线分析结果如图3及表7所示,结果显示,OAS2N检测法和OAS2(IFNa+N-N)检测法的ROC曲线下面积(AUC)分别为0.774、0.814;灵敏度分别为72%、71%;特异性分别为80%、83%。
上述结果表明,2种检测方法均具有较高的灵敏度和特异性,单独使用OAS2N检测法或OAS2(IFNa+N-N)检测法可以用于预测IFNα对CHB患者的治疗效果或CHB患者对IFNα治疗的应答,具有较高的灵敏度和特异性。
表6.OAS2N检测法和OAS2(IFNa+N-N)检测法的主要步骤
表7.OAS2N检测法、OAS2(IFNa+N-N)检测法的ROC分析
实施例5.OAS2(IFNa+N-N)检测法、USP18(IFNa+N-N)检测法及两者联合检测的ROC曲线分析
根据实施例3中OAS2(IFNa+N-N)组和USP18(IFNa+N-N)组的实验条件,分别建立了相应的OAS2(IFNa+N-N)检测法和USP18(IFNa+N-N)检测法,表8记载了上述两种方法的主要步骤。本实施例采用了如上所述的两种方法分别对实施例3中所记载的CHB患者样本进行了检测,分别获得两个诊断指标:OAS2的mRNA相对表达水平和USP18的mRNA相对表达水平;然后使用SPSS 17.0软件以OAS2的mRNA相对表达水平或USP18的mRNA相对表达水平为诊断指标,绘制ROC曲线以检验单独使用OAS2(IFNa+N-N)检测法或USP18(IFNa+N-N)检测法在预测IFNα对CHB患者的治疗效果或CHB患者对IFNα治疗的应答方面的性能;并且将上述指标作为联合检测指标,通过Logistic回归模型,获得统计分析值,以该值再次绘制ROC曲线,评价两者联合检测的预测性能。ROC曲线分析结果
如图4及表9所示,结果显示,2种方法的ROC曲线下面积(AUC)分别为0.814、0.823;灵敏度分别为71%、60%;特异性分别为83%、95%。2种方法联合检测的AUC可以达到0.861,灵敏度和特异性分别为74%、89%。上述结果表明OAS2(IFNa+N-N)检测法和USP18(IFNa+N-N)检测法及联合检测均具有良好的灵敏度和特异性,进一步无论是表明单独使用OAS2N检测法或OAS2(IFNa+N-N)检测法,还是两者联合检测,均对预测IFNα对CHB患者的治疗效果或CHB患者对IFNα治疗的应答具有很好的灵敏度和特异性。
表8.OAS2(IFNa+N-N)检测法和USP18(IFNa+N-N)检测法的主要步骤
表9.OAS2(IFNa+N-N)检测法和USP18(IFNa+N-N)检测法以及两者联合检测的ROC分析
尽管本发明的具体实施方式已经得到详细的描述,但本领域技术人员将理解:根据已经公布的所有教导,可以对细节进行各种修改和变动,并且这些改变均在本发明的保护范围之内。本发明的全部分为由所附权利要求及其任何等同物给出。
Claims (9)
- 一种试剂盒,其包括能够检测标志物表达水平的第一试剂,任选地还包括IFNα和/或刺激物;其中,所述标志物选自OAS2或USP18,所述刺激物选自HBV抗原、HBV抗原的抗原性片段或其任何组合;优选地,所述HBV抗原为HBcAg;更优选地,所述HBcAg具有如SEQ ID NO:38所示的氨基酸序列;优选地,所述抗原性片段具有选自下列的氨基酸序列:SEQ ID NO:3-37;优选地,所述刺激物包含分别具有如SEQ ID NO:3-37所示的氨基酸序列的抗原性片段;优选地,所述第一试剂为能够检测所述标志物的mRNA水平或蛋白水平的试剂,例如为能够检测所述标志物的mRNA水平的试剂。
- 权利要求1所述的试剂盒,其包含能够检测OAS2表达水平的第一试剂,任选地还包括IFNα和/或刺激物。
- 权利要求1所述的试剂盒,其包含能够检测USP18表达水平的第一试剂和IFNα,任选地还包括刺激物。
- 权利要求1-3任一项所述的试剂盒,其还包含用于预处理含有PBMC的样品的第二试剂,其中所述第二试剂包括选自下列的一种或多种试剂:用于稀释样品的稀释剂(例如,磷酸盐缓冲液或生理盐水);用于阻止血液凝固的抗凝剂(例如,肝素);用于维持PBMC活性的试剂(例如,培养基或培养液);和,用于分离PBMC的试剂(例如,淋巴细胞分离液,例如聚蔗糖-泛影葡胺溶液);更优选地,所述第二试剂包括用于维持PBMC活性的试剂(例如,培养基或培养液),和/或用于分离PBMC的试剂(例如,淋巴细胞分离液,例如聚蔗糖-泛影葡胺溶液);优选地,所述试剂盒还包含采血装置(例如无热原真空采血管)和/或PBMC分离装置(例如PBMC细胞分离管)。
- 能够检测标志物表达水平的试剂在制备试剂盒中的用途,所述试剂盒用于预测IFNα对患有慢性乙肝的受试者的治疗效果或所述受试者对IFNα治疗的应答;其中,所述标志物选自OAS2或USP18;优选地,所述试剂盒还包括IFNα和/或刺激物,所述刺激物选自HBV抗原、HBV抗原的抗原性片段或其任何组合;优选地,所述HBV抗原为HBcAg;更优选地,所述HBcAg具有如SEQ ID NO:38所示的氨基酸序列;优选地,所述抗原性片段具有选自下列的氨基酸序列:SEQ ID NO:3-37;优选地,所述刺激物包含分别具有如SEQ ID NO:3-37所示的氨基酸序列的抗原性片段;优选地,所述能够检测标志物表达水平的试剂为能够测定所述标志物的mRNA水平或蛋白水平的试剂,例如为能够测定所述标志物的mRNA水平的试剂;更优选地,所述能够检测标志物表达水平的试剂通过PCR定量测定所述标志物的mRNA水平;优选地,所述试剂盒还包含一种或多种选自1)-6)的试剂或装置:1)用于稀释样品的稀释剂,例如磷酸盐缓冲液或生理盐水;2)用于阻止血液凝固的抗凝剂,例如肝素;3)用于维持PBMC活性的试剂,例如培养基或培养液;4)用于分离PBMC的试剂,例如淋巴细胞分离液,例如聚蔗糖-泛影葡胺溶液;5)采血装置,例如无热原真空采血管;和6)PBMC分离装置,例如PBMC细胞分离管;优选地,所述试剂盒包括用于维持PBMC活性的试剂(例如,培养基或培养液),和/或用于分离PBMC的试剂(例如,淋巴细胞分离液,例如聚蔗糖-泛影葡胺溶液)。
- 权利要求5所述的用途,其中所述试剂盒通过包括下述步骤的方法来预测IFNα对患有慢性乙肝的受试者的治疗效果或所述受试者对IFNα治疗的应答:(1)使用能够检测标志物表达水平的试剂测定来自所述受试者的样品中所述标志物的表达水平,其中所述标志物为OAS2;和(2)将该表达水平与参考值进行比较,或对该表达水平进行统计学分析以获得统计分析值,并将该统计分析值与参考值进行比较,并判断所述受试者是否会对IFNα 治疗产生应答;其中,所述样品包含外周血单个核细胞(PBMC),例如全血(例如抗凝全血)、外周血单个核细胞(PBMC)、或外周血白膜层;优选地,在步骤(2)中,使用Logistic回归模型对所述表达水平进行统计分析;优选地,在步骤(1)之前,所述方法还包括下列步骤中的一项或多项:(a)使用采血装置从所述受试者获得样品;(b)使用抗凝剂处理采血装置或来自所述受试者的样品;(c)使用用于维持PBMC活性的试剂处理来自所述受试者的样品;(d)使用稀释剂稀释来自所述受试者的样品;和(e)使用用于分离PBMC的试剂或PBMC分离装置从来自所述受试者的样品中分离获得PBMC。
- 权利要求5所述的用途,其中所述试剂盒通过包括下述步骤的方法来预测IFNα对患有慢性乙肝的受试者的治疗效果或所述受试者对IFNα治疗的应答:(1)使用IFNα刺激来自所述受试者的至少一份样品作为待测样品,并将未经IFNα刺激的样品作为对照样品;或,使用IFNα和刺激物共同刺激来自所述受试者的至少一份样品作为待测样品,并将经所述刺激物刺激但未经IFNα刺激的样品作为对照样品;其中,所述刺激物选自HBV抗原、HBV抗原的抗原性片段或其任何组合;(2)使用能够检测标志物表达水平的试剂测定步骤(1)中各个样品中所述标志物的表达水平,并获得所述待测样品中该标志物的表达水平相比于所述对照样品中该标志物的表达水平的变化倍数,将该变化倍数作为所述待测样品中所述标志物的相对表达水平,其中所述标志物选自OAS2或USP18;和(3)将该相对表达水平与参考值进行比较,或对该相对表达水平进行统计学分析以获得统计分析值,并将该统计分析值与参考值进行比较,并判断所述受试者是否会对IFNα治疗产生应答;其中,所述样品包含外周血单个核细胞(PBMC),例如全血(例如抗凝全血)、外周血单个核细胞(PBMC)、或外周血白膜层;优选地,在步骤(2)中,通过比较Ct法获得所述待测样品中该标志物的mRNA水平相比于所述对照样品中该标志物的mRNA水平的变化倍数,即所述待测样品中所述标志物的mRNA相对表达水平;优选地,在步骤(3)中,使用Logistic回归模型对所述相对表达水平进行统计 分析;优选地,在步骤(1)之前,所述方法还包括下列步骤中的一项或多项:(a)使用采血装置从所述受试者获得样品;(b)使用抗凝剂处理采血装置或来自所述受试者的样品;(c)使用用于维持PBMC活性的试剂处理来自所述受试者的样品;(d)使用稀释剂稀释来自所述受试者的样品;和(e)使用用于分离PBMC的试剂或PBMC分离装置从来自所述受试者的样品中分离获得PBMC。
- 用于预测IFNα对患有慢性乙肝的受试者的治疗效果或所述受试者对IFNα治疗的应答的方法,或获得诊断分析的结果的方法,其中所述诊断分析用于预测IFNα对患有慢性乙肝的受试者的治疗效果或所述受试者对IFNα治疗的应答,其包括下述步骤:(1)提供来自所述受试者的样品;(2)测定所述样品中标志物的表达水平,其中所述标志物为OAS2;和(3)将该表达水平与参考值进行比较,或对该表达水平进行统计学分析以获得统计分析值,并将该统计分析值与参考值进行比较,并判断所述受试者是否会对IFNα治疗产生应答;其中,所述样品包含外周血单个核细胞(PBMC),例如全血(例如抗凝全血)、外周血单个核细胞(PBMC)、或外周血白膜层;优选地,在步骤(2)中,测定所述标志物的mRNA水平或蛋白水平,例如测定所述标志物的mRNA水平;更优选地,在步骤(2)中,通过PCR定量测定所述标志物的mRNA水平;优选地,在步骤(3)中,使用Logistic回归模型对所述表达水平进行统计分析;优选地,在步骤(1)之前,还包括下列步骤中的一项或多项:(a)从所述受试者获得样品;(b)向来自所述受试者的样品中加入抗凝剂,例如肝素;(c)从来自所述受试者的样品中获取PBMC或含有PBMC的血液成分(例如,外周血白膜层);(d)向来自所述受试者的样品中加入培养液或培养基;和,(e)稀释来自所述受试者的样品。
- 用于预测IFNα对患有慢性乙肝的受试者的治疗效果或所述受试者对IFNα治疗的应答的方法,或获得诊断分析的结果的方法,其中所述诊断分析用于预测IFNα对 患有慢性乙肝的受试者的治疗效果或所述受试者对IFNα治疗的应答,其包括下述步骤:(1)提供来自所述受试者的至少两份样品;(2)使用IFNα刺激至少一份样品作为待测样品,并将未经IFNα刺激的样品作为对照样品;或,使用IFNα和刺激物共同刺激来自所述受试者的至少一份样品作为待测样品,并将经所述刺激物刺激但未经IFNα刺激的样品作为对照样品;其中,所述刺激物选自HBV抗原、HBV抗原的抗原性片段或其任何组合;(3)测定步骤(2)中各个样品中标志物的表达水平,并计算所述待测样品中该标志物的表达水平相比于所述对照样品中该标志物的表达水平的变化倍数,将该变化倍数作为所述待测样品中所述标志物的相对表达水平,其中所述标志物选自OAS2或USP18;和(4)将该相对表达水平与参考值进行比较,或对该相对表达水平进行统计学分析以获得统计分析值,并将该统计分析值与参考值进行比较,并判断所述受试者是否会对IFNα治疗产生应答;其中,所述样品包含外周血单个核细胞(PBMC),例如全血(例如抗凝全血)、外周血单个核细胞(PBMC)、或外周血白膜层;优选地,在步骤(2)中,所述HBV抗原为HBVcAg;更优选地,所述HBVcAg具有如SEQ ID NO:38所示的氨基酸序列;优选地,所述抗原性片段具有选自下列的氨基酸序列:SEQ ID NO:3-37;更优选地,所述刺激物包含分别具有如SEQ ID NO:3-37所示的氨基酸序列的抗原性片段;优选地,在步骤(3)中,测定所述标志物的mRNA水平或蛋白水平;例如测定所述标志物的mRNA水平;更优选地,在步骤(3)中,通过PCR定量测定所述标志物的mRNA水平;优选地,在步骤(3)中,通过比较Ct法获得所述待测样品中该标志物的mRNA水平相比于所述对照样品中该标志物的mRNA水平的变化倍数,即所述待测样品中所述标志物的mRNA相对表达水平;优选地,在步骤(4)中,使用Logistic回归模型对所述相对表达水平进行统计分析;优选地,在步骤(1)之前,还包括下列步骤中的一项或多项:(a)从所述受试者获得样品;(b)向来自所述受试者的样品中加入抗凝剂,例如肝素;(c)从来自 所述受试者的样品中获取PBMC或含有PBMC的血液成分(例如,外周血白膜层);(d)向来自所述受试者的样品中加入培养液或培养基;和,(e)稀释来自所述受试者的样品。
Applications Claiming Priority (2)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| CN201610121177 | 2016-03-04 | ||
| CN201610121177.6 | 2016-03-04 |
Publications (1)
| Publication Number | Publication Date |
|---|---|
| WO2017148432A1 true WO2017148432A1 (zh) | 2017-09-08 |
Family
ID=59743520
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| PCT/CN2017/075542 WO2017148432A1 (zh) | 2016-03-04 | 2017-03-03 | 用于预测慢性乙肝患者对IFNα治疗的应答的方法和试剂盒 |
Country Status (2)
| Country | Link |
|---|---|
| CN (1) | CN107151697B (zh) |
| WO (1) | WO2017148432A1 (zh) |
Cited By (1)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN115679003A (zh) * | 2022-10-28 | 2023-02-03 | 北京大学 | 用于预测药物疗效的组合物、系统及用途 |
Citations (2)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN101821629A (zh) * | 2007-09-10 | 2010-09-01 | 诺瓦提斯研究基金会弗里德里克·米谢尔生物医学研究所 | 用于预测患有肝脏病毒性感染的受试者针对抗病毒疗法的应答的方法 |
| CN104603289A (zh) * | 2012-06-15 | 2015-05-06 | 哈里·斯泰利 | 检测疾病或病状的方法 |
Family Cites Families (1)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN102485907B (zh) * | 2010-12-06 | 2014-03-05 | 北京大学人民医院 | 干扰素治疗慢性丙型肝炎转归预测基因芯片 |
-
2017
- 2017-03-03 WO PCT/CN2017/075542 patent/WO2017148432A1/zh active Application Filing
- 2017-03-03 CN CN201710121980.4A patent/CN107151697B/zh active Active
Patent Citations (2)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN101821629A (zh) * | 2007-09-10 | 2010-09-01 | 诺瓦提斯研究基金会弗里德里克·米谢尔生物医学研究所 | 用于预测患有肝脏病毒性感染的受试者针对抗病毒疗法的应答的方法 |
| CN104603289A (zh) * | 2012-06-15 | 2015-05-06 | 哈里·斯泰利 | 检测疾病或病状的方法 |
Cited By (1)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| CN115679003A (zh) * | 2022-10-28 | 2023-02-03 | 北京大学 | 用于预测药物疗效的组合物、系统及用途 |
Also Published As
| Publication number | Publication date |
|---|---|
| CN107151697A (zh) | 2017-09-12 |
| CN107151697B (zh) | 2021-05-07 |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| KR20160052585A (ko) | 항-tl1a 치료를 위한 시스템, 장치 및 방법 | |
| JP5650871B1 (ja) | そう痒を伴う疾患に罹患した患者のil−31アンタゴニストによる治療に対する応答を予測する方法 | |
| JP2014158500A (ja) | 患者の癌の進行およびその転帰の生体外予後診断方法、および該方法を実施するための手段 | |
| AU2013229762A1 (en) | Biomarker compositions and methods | |
| EP2191274A1 (en) | Method for predicting the response of a subject suffering from a viral infection of the liver to an antiviral therapy | |
| US20210148916A1 (en) | Methods for Monitoring Polymorphonuclear Myeloid Derived Suppressor Cells and Compositions and Methods of Treatment of Cancer | |
| EP2271776A1 (en) | Antiviral therapy | |
| JP2021502069A (ja) | がんを検出するための非コードrna | |
| Rasmussen et al. | Chronic immune activation is a distinguishing feature of liver and PBMC gene signatures from HCV/HIV coinfected patients and may contribute to hepatic fibrogenesis | |
| EP3922641A1 (en) | Methods for detecting and treating covid patients requiring intensive care | |
| JP2021126107A (ja) | 膵癌・胆道系癌の化学療法剤の奏効性予測マーカー、及びそれに対応する奏効性予測キット | |
| US20210293814A1 (en) | Systems and methods for spectral imaging characterization of macrophages for use in personalization of targeted therapies to prevent fibrosis development in patients with chronic liver disease | |
| Hong et al. | Multiomics profiling of buffy coat and plasma unveils etiology-specific signatures in hepatocellular carcinoma | |
| JP2020531439A (ja) | アトピー性皮膚炎の治療及び治療選択のための組成物及び方法 | |
| Li et al. | Recent developments in affinity-based selection of aptamers for binding disease-related protein targets | |
| WO2017148432A1 (zh) | 用于预测慢性乙肝患者对IFNα治疗的应答的方法和试剂盒 | |
| WO2017125007A1 (zh) | 用于诊断活动性结核的方法和试剂盒 | |
| KR101704828B1 (ko) | 체액 내 세포밖 소포체의 단백질 또는 유전자 분석을 통한 염증성 질환 진단 방법 | |
| JP2013021932A (ja) | 関節リウマチに対する抗il−6受容体抗体療法の有効性の予測方法 | |
| JP7554779B2 (ja) | 処置レジメンを調節する方法 | |
| KR101503039B1 (ko) | C형 간염 바이러스 진단용 프라이머, 프로브, 이를 포함하는 키트 및 상기 키트를 이용한 c형 간염 바이러스 진단 방법 | |
| JP2011211970A (ja) | インターフェロンに対する感受性又は応答性を予測する方法、キット及びマーカー遺伝子 | |
| WO2019213619A1 (en) | Hbv diagnostic, prognostic, and therapeutic methods and products | |
| CN114381460B (zh) | 一种用于检测腮腺炎病毒的核酸适配体、试剂盒及应用 | |
| JP4863452B2 (ja) | ヒトTh1/Th2分化誘導系及びその用途 |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| NENP | Non-entry into the national phase |
Ref country code: DE |
|
| 121 | Ep: the epo has been informed by wipo that ep was designated in this application |
Ref document number: 17759283 Country of ref document: EP Kind code of ref document: A1 |
|
| 122 | Ep: pct application non-entry in european phase |
Ref document number: 17759283 Country of ref document: EP Kind code of ref document: A1 |