WO2010103522A1 - Method for detection of nucleic acid sequences - Google Patents
Method for detection of nucleic acid sequences Download PDFInfo
- Publication number
- WO2010103522A1 WO2010103522A1 PCT/IL2010/000206 IL2010000206W WO2010103522A1 WO 2010103522 A1 WO2010103522 A1 WO 2010103522A1 IL 2010000206 W IL2010000206 W IL 2010000206W WO 2010103522 A1 WO2010103522 A1 WO 2010103522A1
- Authority
- WO
- WIPO (PCT)
- Prior art keywords
- nucleic acid
- sequence
- seq
- probe
- primer
- Prior art date
Links
Classifications
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12Q—MEASURING OR TESTING PROCESSES INVOLVING ENZYMES, NUCLEIC ACIDS OR MICROORGANISMS; COMPOSITIONS OR TEST PAPERS THEREFOR; PROCESSES OF PREPARING SUCH COMPOSITIONS; CONDITION-RESPONSIVE CONTROL IN MICROBIOLOGICAL OR ENZYMOLOGICAL PROCESSES
- C12Q1/00—Measuring or testing processes involving enzymes, nucleic acids or microorganisms; Compositions therefor; Processes of preparing such compositions
- C12Q1/68—Measuring or testing processes involving enzymes, nucleic acids or microorganisms; Compositions therefor; Processes of preparing such compositions involving nucleic acids
- C12Q1/6813—Hybridisation assays
Definitions
- the invention relates in general to a method for detecting RNA sequences, particularly but not limited to microRNA and other short RNA sequences.
- miRNAs are an abundant class of short ( ⁇ 22nt) non-coding single- stranded RNAs. They typically bind to complementary sequences at the 3 '-untranslated region (UTR) of mRNAs and thereby exert postranscriptional regulation on gene expression and protein translation.
- UTR 3 '-untranslated region
- miRNAs play critical roles in many key biological processes, such as cell growth, tissue differentiation, cell proliferation, embryonic development, apoptosis, viral infection, cancer and more.
- miRNAs expression is tissue specific so much so that they were suggested to serve as biomarkers and to accurately identify cancers of unknown tissue origin.
- RTPCR Real time PCR
- stem-loop primers are used to reverse transcribe (RT) the miRNA, and the RT product is quantified in a TaqManTM PCR (qRTPCR) assay) (Chen, C. Nucleic Acids Research, Vol. 33, No. 20, 2005).
- qRTPCR TaqManTM PCR
- Varkonyi-Gasic et al ⁇ Plant Methods 3:12, 2007 have detected and quantified miRNAs by stem loop reverse primer and qRTPCR using a generic and small universal probe.
- the Varkonyi-Gasic method is based on the method first described by Chen et al., with replacement of the specific probe by a generic probe.
- This one step method also requires a unique stem-loop-specific reverse transcription primer for each miRNA in addition to the miRNA specific forward primer.
- This generic primer is 50nts long, and thus requires a special synthesis scale. Furthermore, amplification is limited to 35 cycles due to non-specific amplification in higher cycles.
- SYBR Green intercalates into double-stranded DNA in a non-specific manner, it commonly detects non-specific PCR products and primer-dimers, leading to false-positive results. Due to the small size of these products, in most cases melting curve analysis and gel electrophoresis are unable to distinguish between the specific and non-specific PCR products. For these reasons the PCR-product can not be quantified directly.
- RNA particularly mRNA and miRNA.
- the present invention provides a method of detecting a target RNA nucleic acid sequence in a biological sample, the method comprising: providing the biological sample comprising the target RNA nucleic acid sequence, annealing the target RNA nucleic acid sequence with a poly(T) primer comprising a 5' universal adaptor sequence, generating a reverse transcript of the polyadenylated RNA, with the poly(T) primer, and amplifying the reverse transcript product by a polymerase chain reaction (PCR) comprising a specific forward primer, a universal reverse primer and a general probe, to generate an amplicon; wherein the forward primer is at least partially identical to the target RNA nucleic acid sequence and the universal reverse primer is at least partially identical to a 5' region of the adaptor sequence of the poly (T) primer.
- PCR polymerase chain reaction
- the target RNA nucleic acid sequence is a short nucleic acid, and it is extended at the 3' end by polyadenylation, prior to its annealing with the poly(T) primer.
- the present invention further provides a kit for the detection of a target RNA nucleic acid sequence in a biological sample, the kit comprising a poly(T) primer comprising a 5 1 universal adaptor sequence, a specific forward primer, a universal reverse primer and a general probe, wherein the forward primer is at least partially identical to the target RNA nucleic acid sequence, and the universal reverse primer is at least partially identical to a 5' region of the adaptor sequence of the poly (T) primer.
- the general probe is partially complementary to the adaptor sequence region of the sense strand of the amplicon. According to some embodiments the general probe is partially further partially complementary to the poly(T) region of the sense strand of the amplicon. According to additional embodiments the general probe is complementary to the adaptor sequence region of the antisense strand of the amplicon. According to some embodiments the general probe is further partially complementary to the poly(T) region of the antisense strand of the amplicon. According to some embodiments the general probe comprises a sequence selected from the, group consisting of SEQ. ID NOS: 48-50. According to some embodiments the general probe which is partially complementary to the adaptor sequence region of the sense strand of the amplicon comprises a sequence selected from the group consisting of SEQ.
- the general probe which is further partially complementary to the poly(T) region of the sense strand of the amplicon comprises SEQ. ID. NO: 48.
- the general probe which is complementary to the adaptor sequence region of the antisense strand of the amplicon, and further partially complementary to the poly(T) region of the antisense strand of the amplicon comprises SEQ. ID. NO: 49.
- the forward primer comprises a sequence selected from the group consisting of SEQ. ID NOS. 2, 4, 6, 8, 12, 14, 16, 18, 20, 22, 24, 40-47, 52, 55, 57, 58, 63, 64, 69-87, 89 and 91.
- the reverse primer comprises the sequence of SEQ. ID NO. 28.
- the method of the invention comprises SEQ. ID NO. 60 as a normalizer.
- the target RNA nucleic acid sequence is selected from the group consisting of: mRNA, microRNA, siRNA, PiwiRNA, and a combination thereof. According to some embodiments the target RNA nucleic acid sequence is a short RNA nucleic acid sequence selected from the group consisting of: microRNA, siRNA, PiwiRNA, and a combination thereof. According to some embodiments the target RNA nucleic acid sequence is a short RNA nucleic acid sequence 17-25 nt in length. According to some embodiments the general probe comprises a fluorescent reporter group and a quencher. The general probe may comprise a 5' fluorescent reporter group and 3' a quencher.
- the general probe may comprise a 3' fluorescent reporter group and a 5' quencher.
- the general probe is a TaqMan probe.
- the general probe further comprises a minor groove binder (MGB).
- the general probe further comprises locked nucleic acids (LNA).
- the biological sample is selected from the group consisting of bodily fluid, a cell line, a tissue sample, a biopsy sample, a needle biopsy sample, a surgically removed sample, and a sample obtained by tissue-sampling procedures.
- the bodily fluid is serum.
- the tissue is a fresh, frozen, fixed, wax-embedded or formalin-fixed paraffin- embedded (FFPE) tissue.
- FFPE formalin-fixed paraffin- embedded
- Figure IA Schematic description of the General (antisense) Probe Detection Assay of the invention.
- Step 1 (top vertical arrow): polyadenylation of total RNA to generate poly(A) miRNA.
- Step 2 reverse transcription of poly(A) miRNA using a poly(T) primer that contains a universal tail (adaptor) and a probe binding region.
- Step 3 Real Time PCR amplification using a miRNA-specific forward primer, a reverse primer complementary to the universal tail and a general antisense probe.
- FIG. 1B Schematic description of the Varkonyi-Gasic (stem loop) assay.
- Step 1 top vertical arrow: stem-loop RT primers bind to the 3' portion of the miRNA and are reverse transcribed;
- Step 2 bottom vertical arrow: PCR amplification using miRNA-specific forward primer, specific reverse primer and a TaqMan probe.
- Figures 2A-2D present amplification curves, in which the y-axis represents fluorescence (483-533) and the x-axis represents the number of cycles.
- the amounts of cDNA/RNA used for each reaction were as follows: (1) 2.5 ng; (2) 250 pg; (3) 25 pg; (4) 2.5 pg; (5) 0.25 pg; (-RT, no reverse transcriptase negative control) 2.5 ng RNA; (NTC) no- template negative control.
- NTC no- template negative control.
- FIG. 3A SYBR-Green qPCR amplification of hsa-miR-296-3p (SEQ. ID NO: 11). The arrow indicates the location of the band of the mature miR.
- Fig. 3B Varkonyi-Gasic stem-loop qPCR amplification of hsa-miR-181a (SEQ. ID NO: 7).
- the arrow indicates the location of the band of the mature miR.
- Fig. 3C General (antisense) Probe amplification: left- hsa-miR-181a (SEQ. ID NO: 7); right- hsa-miR-296-3p (SEQ. ID NO: 11).
- the arrow indicates the location of a band of
- Fig. 4A Correlation of B95-8 cDNA input to the AO-Cp value of the reaction (mean of three experiments).
- the x-axis represents B95-8 RNA (pg/reaction) and the y-axis AQ-Cp.
- the x-axis represents the number of cycles, and the y-axis - fluorescence (483-533).
- B95-8 cDNA was diluted two folds from 250 pg (no UKF-cDNA) to 0.244 pg, in UKF-NB4-CDNA. Zero is UKF-cDNA only (no B95-8 cDNA).
- Fig. 5A Detection of various hsa-miRNAs using the General (antisense) Probe Detection Assay. Correlation of cDNA input (x-axis, cDNA libraries prepared from UKF-NB4 total RNA, pg/reaction) to the 40-Cp values (y-axis) for five host miRNAs.
- hsa-miR-30a SEQ. ID NO: l
- hsa-miR-103 SEQ. ID NO: 3
- black square symbols hsa-miR- 107 (SEQ. ID NO: 5) - white square symbols;
- hsa-miR-181a SEQ. ID NO: 7 - triangles; hsa-miR-210 (SEQ. ID NO: 9) - circles.
- Fig. 5B Correlation between B95-8 cDNA (pg/reaction) input (x-axis), and AQ-Cp values (y- axis) of two EBV-miRNAs [BART-3 (SEQ. ID NO: 33) and BHRF-1-2 (SEQ. ID NO: 35), black diamonds and squares, respectively] and their matching miRNAs* [BART-3* (SEQ. ID NO: 34) and BHRF-1-2* (SEQ. ID NO: 36), open black diamonds and squares, respectively], using cDNA libraries prepared from l ⁇ g of total RNA extracted from B95-8 cells.
- Figure 6 Expression of pattern of various EBV miRNAs in PBMCs in vivo.
- the x-axis depicts the EBV-miRNAs tested and the y-axis depicts the 75-Q? value for each sample.
- the General Probe Detection Assay for EBV-miRNAs was performed on RNAs extracted from ⁇ 10 7 PBMC/sample of two healthy carriers (white and grey bars) and an infectious mononucleosis patient (black bar); the no-template-controls for each of the EBV-miRs was negative.
- Figure 7 Analysis of qPCR amplification on 2,5% agarose gel.
- the left arrow marks the lane with the cDNA including cre-miR1142 (SEQ. ID NO: 51); the right arrow marks the lane with water as a control.
- Figure 8 The sensitivity of the General (sense) Probe Detection Assay: a competition assay demonstrating the assays ability to detect specific miRs in the presence of others.
- Fig 8A Detection of liver-specific hsa-miR-122 (SEQ. ID NO: 61) in mixture of cDNA of total RNA from Liver and Brain tissues.
- the y-axis depicts the PCR signal normalized by subtracting it from 55 (55-Ct); the x-axis depicts the percentage of liver cDNA in the mixture of cDNA of total RNA from Liver and Brain tissues.
- Fig 8B Detection of brain-specific hsa-miR-124 (SEQ. ID NO: 62) in mixture of cDNA of total RNA from Liver and Brain tissues.
- the y-axis depicts the PCR signal normalized by subtracting it from 55 (55-Ct); the x-axis depicts the percentage of brain cDNA in the mixture of cDNA of total RNA from Liver and Brain tissues.
- the present invention provides a sensitive, specific and accurate method for the detection of RNA, including mRNAs and short RNAs such as miRNAs.
- RNA including mRNAs and short RNAs such as miRNAs.
- the RT and the reverse primers, as well as the probe are generic, and the only specific component is the forward primer which determines the amplification of a specific RNA.
- the General Probe Detection assay is able to detect various viral, plant and human miRNAs in several cell lines with a very high sensitivity, and can also discriminate miRNAs that differ by as little as a single nucleotide, exhibiting high specificity.
- preparation of cDNA library and qRT-PCR by this method takes less than one day. This approach enables a versatile and cost effective detection of RNAs. Thus, this protocol can be applied to sensitive, robust, and rapid detection of miRNAs from various origins.
- each intervening number there between with the same degree of precision is explicitly contemplated.
- the numbers 7 and 8 are contemplated in addition to 6 and 9, and for the range 6.0-7.0, the number 6.0, 6.1, 6.2, 6.3, 6.4, 6.5, 6.6, 6.7, 6.8, 6.9 and 7.0 are explicitly contemplated.
- adaptors are oligonucleotides that are continuous to a cDNA of a target nucleic acid, such that PCR primers may anneal (entirely or partially) thereto for amplification of the target nucleic acid.
- Adaptor sequences may also be referred to as tail sequences.
- amplicons are fragments of DNA formed as the products of natural or artificial amplification. antisense
- antisense refers to nucleotide sequences which are reverse complementary to a specific DNA or RNA sequence.
- antisense strand is used in reference to a nucleic acid strand that is complementary to the "sense" strand.
- Antisense molecules may be produced by any method, including synthesis by ligating the gene(s) of interest in a reverse orientation to a viral promoter which permits the synthesis of a complementary strand. Once introduced into a cell, this transcribed strand combines with natural sequences produced by the cell to form duplexes. These duplexes then block either the further transcription or translation. In this manner, mutant phenotypes may be generated. antisense probe
- antisense probe refers to a probe which anneals to the cDNA polyadenylated strand (+ strand) of the amplicon comprising the sequence to be detected by the probe.
- sequence of the antisense probe of the invention may comprise the sequences of SEQ. ID NOS: 48 and 50. attached
- “Attached” or “immobilized” as used herein refer to a probe and a solid support and may mean that the binding between the probe and the solid support is sufficient to be stable under conditions of binding, washing, analysis, and removal.
- the binding may be covalent or non-covalent. Covalent bonds may be formed directly between the probe and the solid support or may be formed by a cross linker or by inclusion of a specific reactive group on either the solid support or the probe, or both.
- Non-covalent binding may be one or more of electrostatic, hydrophilic, and hydrophobic interactions. Included in non-covalent binding is the covalent attachment of a molecule, such as streptavidin, to the support and the non- covalent binding of a biotinylated probe to the streptavidin. Immobilization may also involve a combination of covalent and non-covalent interactions.
- biological sample such as streptavidin
- Bio sample as used herein means a sample of biological tissue or fluid that comprises nucleic acids. Samples may originate from viruses, plants or animals including humans. Such samples include, but are not limited to, tissue or fluid isolated from subjects.
- Biological samples may also include sections of tissues such as biopsy and autopsy samples, formalin-fixed paraffin-embedded (FFPE) samples, frozen sections taken for histological purposes, blood, plasma, serum, sputum, stool, tears, mucus, hair, and skin.
- Biological samples also include explants and primary and/or transformed cell cultures derived from animal or patient tissues .
- Biological samples may also be blood, a blood fraction, urine, effusions, ascitic fluid, saliva, cerebrospinal fluid, cervical secretions, vaginal secretions, endometrial secretions, gastrointestinal secretions, bronchial secretions, sputum, cell line, tissue sample, cellular content of fine needle aspiration (FNA) or secretions from the breast.
- a biological sample may be provided by removing a sample of cells from a subject, but can also be accomplished by using previously isolated cells (e.g., isolated by another person, at another time, and/or for another purpose), or by performing the methods described herein in vivo.
- Archival tissues such as those having treatment or outcome history, may also be used.
- complement "Complement” or “complementary” as used herein means Watson-Crick (e.g., A-
- a full complement or fully complementary may mean 100% complementary base pairing between nucleotides or nucleotide analogs of nucleic acid molecules. Partial complementary may mean less than 100% complementarity, for example 80% complementarity. In some embodiments, the complementary sequence has a reverse orientation (5 '-3').
- Cycle Threshold of qRT-PCR which is the fractional cycle number at which the fluorescence crosses the threshold. Cp is the crossing point.
- Detection means detecting the presence of a component in a sample. Detection also means detecting the absence of a component. Detection also means measuring the level of a component, either quantitatively or qualitatively. differential expression
- differential expression means qualitative or quantitative differences in the temporal and/or cellular gene expression patterns within and among cells and tissue.
- a differentially expressed gene may qualitatively have its expression altered, including an activation or inactivation, in, e.g., normal versus disease tissue. Genes may be turned on or turned off in a particular state, relative to another state thus permitting comparison of two or more states.
- a qualitatively regulated gene may exhibit an expression pattern within a state or cell type which may be detectable by standard techniques. Some genes may be expressed in one state or cell type, but not in both.
- the difference in expression may be quantitative, e.g., in that expression is modulated, either up-regulated- resulting in an increased amount of transcript, or down-regulated- resulting in a decreased amount of transcript.
- the degree to which expression differs need only be large enough to quantify via standard characterization techniques such as expression arrays, quantitative reverse transcriptase PCR, northern analysis, real-time PCR, in situ hybridization and RNase protection. dynamic range
- Dynamic range as used herein is the ratio between the smallest and largest possible values of a changeable quantity.
- expression profile "Expression profile” as used herein may mean a genomic expression profile, e.g., an expression profile of microRNAs. Profiles may be generated by any convenient means for determining a level of a nucleic acid sequence e.g. quantitative hybridization of microRNA, labeled microRNA, amplified microRNA, cRNA, etc., quantitative PCR, ELISA for quantitation, and the like, and allow the analysis of differential gene expression between two samples.
- a subject or patient tumor sample e.g., cells or collections thereof, e.g., tissues, is assayed. Samples are collected by any convenient method, as known in the art.
- Nucleic acid sequences of interest are nucleic acid sequences that are found to be predictive, including the nucleic acid sequences provided above, where the expression profile may include expression data for 5, 10, 20, 25, 50, 100 or more of, including all of the listed nucleic acid sequences.
- expression profile may also mean measuring the abundance of the nucleic acid sequences in the measured samples. expression ratio
- “Expression ratio” as used herein refers to relative expression levels of two or more nucleic acids as determined by detecting the relative expression levels of the corresponding nucleic acids in a biological sample.
- Fram is used herein to indicate a non-full length part of a nucleic acid or polypeptide.
- a fragment is itself also a nucleic acid or polypeptide, respectively.
- forward primer is the sense direction primer complementary to the antisense strand of the amplicon.
- the forward primer may include additional nucleotides at the 5' end, that are not complement to the target sequence, thereby raising the Tm of the annealing of the primer to the target sequence.
- the sequence of the forward primers of the invention may comprise SEQ ED NOS: 2, 4, 6, 8, 12, 14, 16, 18, 20, 22, 40-47, 52, 55, 57, 58, 63, 64 and 69-87. gene
- Gene as used herein may be a natural (e.g., genomic) or synthetic gene comprising transcriptional and/or translational regulatory sequences and/or a coding region and/or non- translated sequences (e.g., introns, 5'- and 3 '-untranslated sequences).
- the coding region of a gene may be a nucleotide sequence coding for an amino acid sequence or a functional RNA, such as tRNA, rRNA, catalytic RNA, siRNA, miRNA or antisense RNA.
- a gene may also be an niRNA or cDNA corresponding to the coding regions (e.g., exons and miRNA) optionally comprising 5'- or 3 '-untranslated sequences linked thereto.
- a gene may also be an amplified nucleic acid molecule produced in vitro comprising all or a part of the coding region and/or 5'- or 3 '-untranslated sequences linked thereto.
- general probe also: universal probe,
- a general probe binds to common sequence located in all target sequences, unspecific to the detected sequence. This is opposed to a specific probe.
- a general probe may be used in the detection of multiple target sequences.
- the sequence of the general probes of the invention may comprise SEQ. ID NOS: 48-50.
- groove binder/minor groove binder (MGB) groove binder/minor groove binder
- Minor groove binder and/or “minor groove binder” may be used interchangeably and refer to small molecules that fit into the minor groove of double-stranded DNA.
- Minor groove binders may be long, flat molecules that can adopt a crescent-like shape and thus, fit snugly into the minor groove of a double helix, often displacing water.
- Minor groove binding molecules may typically comprise several aromatic rings connected by bonds with torsional freedom such as furan, benzene, or pyrrole rings.
- Minor groove binders may be antibiotics such as netropsin, distamycin, berenil, pentamidine and other aromatic diamidines, Hoechst 33258, SN 6999, aureolic anti-tumor drugs such as chromomycin and mithramycin, CC-1065, dihydrocyclopyrroloindole tripeptide (DPI 3 ), l,2-dihydro-(3H)- pyrrolo[3,2-e]indole-7-carboxylate (CDPI 3 ), and related compounds and analogues, including those described in Nucleic Acids in Chemistry and Biology, 2d ed., Blackburn and Gait, eds., Oxford University Press, 1996, and PCT Published Application No.
- antibiotics such as netropsin, distamycin, berenil, pentamidine and other aromatic diamidines, Hoechst 33258, SN 6999, aureolic anti-tumor drugs such as chrom
- a minor groove binder may be a component of a primer, a probe, a hybridization tag complement, or combinations thereof. Minor groove binders may increase the T m of the primer or a probe to which they are attached, allowing such primers or probes to effectively hybridize at higher temperatures. identity
- Identity or “identity” as used herein in the context of two or more nucleic acids or polypeptide sequences mean that the sequences have a specified percentage of residues that are the same over a specified region. The percentage may be calculated by optimally aligning the two sequences, comparing the two sequences over the specified region, determining the number of positions at which the identical residue occurs in both sequences to yield the number of matched positions, dividing the number of matched positions by the total number of positions in the specified region, and multiplying the result by 100 to yield the percentage of sequence identity.
- the residues of the single sequence are included in the denominator but not the numerator of the calculation.
- thymine (T) and uracil (U) may be considered equivalent.
- Identity may be performed manually or by using a computer sequence algorithm such as BLAST or BLAST 2.0.
- “Inhibit” as used herein may mean prevent, suppress, repress, reduce or eliminate.
- isolated when used in reference to nucleic acids, is intended to mean that a nucleic acid molecule is present in a form that is substantially separated from other naturally occurring nucleic acids that are normally associated with the molecule. Specifically, since a naturally existing chromosome (or a viral equivalent thereof) includes a long nucleic acid sequence, an "isolated nucleic acid” as used herein means a nucleic acid molecule having only a portion of the nucleic acid sequence in the chromosome but not one or more other portions present on the same chromosome.
- an "isolated nucleic acid” typically includes no more than 25 kb naturally occurring nucleic acid sequences which immediately flank the nucleic acid in the naturally existing chromosome (or a viral equivalent thereof).
- an "isolated nucleic acid” as used herein is distinct from a clone in a conventional library such as genomic DNA library and cDNA library in that the clone in a library is still in admixture with almost all the other nucleic acids of a chromosome or cell.
- an "isolated nucleic acid” as used herein also should be substantially separated from other naturally occurring nucleic acids that are on a different chromosome of the same organism.
- an "isolated nucleic acid” means a composition in which the specified nucleic acid molecule is significantly enriched so as to constitute at least 10% of the total nucleic acids in the composition.
- an "isolated nucleic acid” can be a hybrid nucleic acid having the specified nucleic acid molecule covalently linked to one or more nucleic acid molecules that are not the nucleic acids naturally flanking the specified nucleic acid.
- an isolated nucleic acid can be in a vector.
- the specified nucleic acid may have a nucleotide sequence that is identical to a naturally occurring nucleic acid or a modified form or mutant thereof having one or more mutations such as nucleotide substitution, deletion/insertion, inversion, and the like.
- Label as used herein means a composition detectable by spectroscopic, photochemical, biochemical, immunochemical, chemical, or other physical means.
- useful labels include 32 P, fluorescent dyes, electron-dense reagents, enzymes (e.g., as commonly used in an ELISA), biotin, digoxigenin, or haptens and other entities which can be made detectable.
- a label may be incorporated into nucleic acids and proteins at any position. locked nucleic acid (LNA)
- Locked nucleic acids as referred to herein, are modified RNA nucleotides, used to increase the sensitivity and specificity of expression in DNA molecular biology techniques based on oligonucleotides.
- the ribose moiety of an LNA nucleotide is modified with an extra bridge connecting the 2' oxygen and 4' carbon. The bridge "locks" the ribose in the 3'- endo (North) conformation, which is often found in the A-form of DNA or RNA.
- LNA nucleotides can be mixed with DNA or KNA bases in the oligonucleotide when desired.
- the locked ribose conformation enhances base stacking and backbone pre-organization, thereby increasing the thermal stability (melting temperature) of oligonucleotides. mismatch
- “Mismatch” means a nucleobase of a first nucleic acid that is not capable of pairing with a nucleobase at a corresponding position of a second nucleic acid.
- modulation means a perturbation of function or activity, hi certain embodiments, modulation means an increase in gene expression, m certain embodiments, modulation means a decrease in gene expression.
- nucleic acid Nucleic acid or “oligonucleotide” or “polynucleotide” as used herein mean at least two nucleotides covalently linked together. The depiction of a single strand also defines the sequence of the complementary strand.
- a nucleic acid also encompasses the complementary strand of a depicted single strand. Many variants of a nucleic acid may be used for the same purpose as a given nucleic acid. Thus, a nucleic acid also encompasses substantially identical nucleic acids and complements thereof.
- a single strand provides a probe that may hybridize to a target sequence under stringent hybridization conditions.
- a nucleic acid also encompasses a probe that hybridizes under stringent hybridization conditions.
- Nucleic acids may be single stranded or double stranded, or may contain portions of both double stranded and single stranded sequence.
- the nucleic acid may be DNA, both genomic and cDNA, RNA, or a hybrid, where the nucleic acid may contain combinations of deoxyribo- and ribo-nucleotides, and combinations of bases including uracil, adenine, thymine, cytosine, guanine, inosine, xanthine hypoxanthine, isocytosine and isoguanine.
- Nucleic acids may be obtained by chemical synthesis methods or by recombinant methods.
- a nucleic acid will generally contain phosphodiester bonds, although nucleic acid analogs may be included that may have at least one different linkage, e.g., phosphoramidate, phosphorothioate, phosphorodithioate, or O-methylphosphoroamidite linkages and peptide nucleic acid backbones and linkages.
- Other analog nucleic acids include those with positive backbones; non-ionic backbones, and non-ribose backbones, including those described in U.S. Pat. Nos. 5,235,033 and 5,034,506, which are incorporated by reference.
- Nucleic acids containing one or more non-naturally occurring or modified nucleotides are also included within one definition of nucleic acids.
- the modified nucleotide analog may be located for example at the 5'-end and/or the 3'-end of the nucleic acid molecule.
- Representative examples of nucleotide analogs may be selected from sugar- or backbone-modified ribonucleotides. It should be noted, however, that also nucleobase-modified ribonucleotides, i.e. ribonucleotides, containing a non-naturally occurring nucleobase instead of a naturally occurring nucleobase such as uridines or cytidines modified at the 5-position, e.g.
- the 2'-OH-group may be replaced by a group selected from H 5 OR, R, halo, SH, SR, NH 2 , NHR, NR 2 or CN, wherein R is C 1 -C 6 alkyl, alkenyl or alkynyl and halo is F 5 Cl, Br or I.
- Modified nucleotides also include nucleotides conjugated with cholesterol through, e.g., a hydroxyprolinol linkage as described in Krutzfeldt et al, Nature 438:685-689 (2005) and Soutschek et al., Nature 432:173-178 (2004), which are incorporated herein by reference.
- Modifications of the ribose-phosphate backbone may be done for a variety of reasons, e.g., to increase the stability and half-life of such molecules in physiological environments, to enhance diffusion across cell membranes, or as probes on a biochip.
- the backbone modification may also enhance resistance to degradation, such as in the harsh endocytic environment of cells.
- the backbone modification may also reduce nucleic acid clearance by hepatocytes, such as in the liver.
- Mixtures of naturally occurring nucleic acids and analogs may be made; alternatively, mixtures of different nucleic acid analogs, and mixtures of naturally occurring nucleic acids and analogs may be made. partially identical
- partially identical means that a first sequence is at least 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 97%, 98% or 99% identical to a second sequence over a region of 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70, 75, 80, 85, 90, 95, 100 or more nucleotides, or that the two sequences hybridize under stringent hybridization conditions.
- Polyadenylation is the addition of a poly(A) tail to an RNA molecule.
- the poly(A) tail is a stretch of RNA consisting of multiple adenosine monophosphates.
- a primer is a strand of nucleic acid that serves as a starting point for DNA replication. They are required because DNA polymerases can only add new nucleotides to an existing strand of DNA. The polymerase starts replication at the 3'-end of the primer, and copies the opposite strand.
- DNA primers are usually short, chemically synthesized oligonucleotides. They are hybridized to a target DNA, which is then copied by the polymerase.
- a primer-dimer as used herein means a potential by-product in PCR.
- Primer-dimers consist of primer molecules that have hybridized to each other due to complementary bases in the primers.
- the DNA polymerase amplifies the primer-dimer, leading to competition for PCR reagents, thus potentially inhibiting amplification of the DNA sequence targeted for PCR amplification.
- primer-dimer s may interfere with accurate quantification.
- Probe as used herein means an oligonucleotide capable of binding to a target nucleic acid of complementary sequence through one or more types of chemical bonds, usually through complementary base pairing, usually through hydrogen bond formation.
- Probes may bind target sequences lacking complete complementarity with the probe sequence depending upon the stringency of the hybridization conditions. There may be any number of base pair mismatches which will interfere with hybridization between the target sequence and the single stranded nucleic acids described herein. However, if the number of mismatches is so great that no hybridization can occur under even the least stringent of hybridization conditions, the sequence is not a complementary target sequence.
- a probe may be single stranded or partially single and partially double stranded. The strandedness of the probe is dictated by the structure, composition, and properties of the target sequence.
- Probes may be directly labeled or indirectly labeled.
- An exemplary probe is TaqMan MGB probe that contains 5' fluorescence, a 3' quencher arid MGB. promoter
- Promoter means a synthetic or naturally-derived molecule which is capable of conferring, activating or enhancing expression of a nucleic acid in a cell.
- a promoter may comprise one or more specific transcriptional regulatory sequences to further enhance expression and/or to alter the spatial expression and/or temporal expression of same.
- a promoter may also comprise distal enhancer or repressor elements, which can be located as much as several thousand base pairs from the start site of transcription.
- a promoter may be derived from sources including viral, bacterial, fungal, plants, insects, and animals.
- a promoter may regulate the expression of a gene component constitutively, or differentially with respect to cell, the tissue or organ in which expression occurs or, with respect to the developmental stage at which expression occurs, or in response to external stimuli such as physiological stresses, pathogens, metal ions, or inducing agents.
- promoters include the bacteriophage T7 promoter, bacteriophage T3 promoter, SP6 promoter, lac operator-promoter, tac promoter, SV40 late promoter, SV40 early promoter, RSV-LTR promoter, CMV IE promoter, SV40 early promoter or S V40 late promoter and the CMV IE promoter.
- a label may also be a quencher molecule, which when in proximity to another label, may decrease the amount of detectable signal of the other label, such as described in U.S. Patent No. 6,541,618, the contents of which are incorporated herein by reference.
- a label may be incorporated into nucleic acids and proteins at any position.
- reverse primer As used herein, the primer complementary to a sense polyadenylated strand is designated the reverse primer.
- the sequence of the reverse primer of the invention may comprise SEQ. ID NO: 28. selectable marker
- Selectable marker as used herein means any gene which confers a phenotype on a host cell in which it is expressed to facilitate the identification and/or selection of cells which are transfected or transformed with a genetic construct.
- selectable markers include the ampicillin-resistance gene (Amp 1 ), tetracycline-resistance gene (Tc 1 ), bacterial kanamycin-resistance gene (Kan 1 ), zeocin resistance gene, the AURI-C gene which confers resistance to the antibiotic aureobasidin A, phosphinothricin-resistance gene, neomycin phosphotransferase gene (nptU), hygromycin-resistance gene, beta- glucuronidase (GUS) gene, chloramphenicol acetyltransferase (CAT) gene, green fluorescent protein (GFP)-encoding gene and luciferase gene.
- sense probe Amp 1
- Tc 1 tetra
- sense probe refers to a probe which is complementary and hybridizes to the universal region on the antisense strand (- strand) of the amplicon generated by RT-PCR.
- sequence of the sense probe of the invention may comprise the sequence of SEQ. ID NO: 49. stringent hybridization conditions
- Stringent hybridization conditions as used herein mean conditions under which a first nucleic acid sequence (e.g., probe) will hybridize to a second nucleic acid sequence
- Stringent conditions are sequence-dependent and will be different in different circumstances. Stringent conditions may be selected to be about 5-1O 0 C lower than the thermal melting point (T m ) for the specific sequence at a defined ionic strength pH.
- the T m may be the temperature (under defined ionic strength, pH, and nucleic acid concentration) at which 50% of the probes complementary to the target hybridize to the target sequence at equilibrium (as the target sequences are present in excess, at T m , 50% of the probes are occupied at equilibrium).
- Stringent conditions may be those in which the salt concentration is less than about
- 1.0 M sodium ion such as about 0.01-1.0 M sodium ion concentration (or other salts) at pH
- 7.0 to 8.3 and the temperature is at least about 3O 0 C for short probes (e.g., about 10-50 nucleotides) and at least about 6O 0 C for long probes (e.g., greater than about 50 nucleotides).
- Stringent conditions may also be achieved with the addition of destabilizing agents such as formamide.
- a positive signal may be at least 2 to 10 times background hybridization.
- Exemplary stringent hybridization conditions include the following: 50% formamide, 5x SSC, and 1% SDS, incubating at
- Substantially complementary as used herein means that a first sequence is at least 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 97%, 98% or 99% identical to the complement of a second sequence over a region of 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70, 75, 80, 85, 90, 95, 100 or more nucleotides, or that the two sequences hybridize under stringent hybridization conditions.
- substantially identical means that a first and a second sequence are at least 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 97%, 98% or 99% identical over a region of 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70, 75, 80, 85, 90, 95, 100 or more nucleotides or amino acids, or with respect to nucleic acids, if the first sequence is substantially complementary to the complement of the second sequence.
- target nucleic acid is substantially complementary to the complement of the second sequence.
- Target nucleic acid as used herein means a nucleic acid or variant thereof that may be bound by another nucleic acid.
- a target nucleic acid may be a DNA sequence.
- the target nucleic acid may be RNA.
- the target nucleic acid may comprise an rnRNA, tRNA, shRNA, siRNA or Piwi-interacting RNA, or a pri-miRNA, pre-miRNA, miRNA, or anti-miRNA.
- One or more probes may bind the target nucleic acid.
- the target binding site may comprise 5-100 or 10-60 nucleotides.
- the target binding site may comprise a total of 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15,16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30-40, 40- 50, 50-60, 61, 62 or 63 nucleotides.
- tissue sample is tissue obtained from a tissue biopsy using methods well known to those of ordinary skill in the related medical arts.
- the phrase "suspected of being cancerous" as used herein means a cancer tissue sample believed by one of ordinary skill in the medical arts to contain cancerous cells. Methods for obtaining the sample from the biopsy include gross apportioning of a mass, microdissection, laser-based microdissection, or other art-known cell-separation methods. universal adaptor
- a universal adaptor is not specific to the amplified target.
- a universal primer is not unique to a specific sequence targeted for amplification, and may hybridize to all sequences comprising a common region complementary to the primer. This is opposed to a specific primer which is specific to the amplified sequence.
- a universal primer may be used in the amplification of multiple target sequences.
- variants as used herein referring to a nucleic acid means (i) a portion of a referenced nucleotide sequence; (ii) the complement of a referenced nucleotide sequence or portion thereof; (iii) a nucleic acid that is substantially identical to a referenced nucleic acid or the complement thereof; or (iv) a nucleic acid that hybridizes under stringent conditions to the referenced nucleic acid, complement thereof, or a sequence substantially identical thereto.
- Vector as used herein means a nucleic acid sequence containing an origin of replication.
- a vector may be a plasmid, bacteriophage, bacterial artificial chromosome or yeast artificial chromosome.
- a vector may be a DNA or RNA vector.
- a vector may be either a self-replicating extrachromosomal vector or a vector which integrates into a host genome.
- wild type As used herein, the term "wild type" sequence refers to a coding, a non-coding or an interface sequence which is an allelic form of sequence that performs the natural or normal function for that sequence. Wild type sequences include multiple allelic forms of a cognate sequence, for example, multiple alleles of a wild type sequence may encode silent or conservative changes to the protein sequence that a coding sequence encodes.
- a gene coding for a microRNA may be transcribed leading to production of an miRNA precursor known as the pri-miRNA.
- the pri-miRNA may be part of a polycistronic RNA comprising multiple pri-miRNAs.
- the pri-miRNA may form a hairpin structure with a stem and loop.
- the stem may comprise mismatched bases.
- the hairpin structure of the pri-miRNA may be recognized by Drosha, which is an RNase III endonuclease. Drosha may recognize terminal loops in the pri-miRNA and cleave approximately two helical turns into the stem to produce a 60-70 nucleotide precursor known as the pre-miRNA. Drosha may cleave the pri-miRNA with a staggered cut typical of RNase III endonucleases yielding a pre-miRNA stem loop with a 5' phosphate and ⁇ 2 nucleotide 3' overhang. Approximately one helical turn of the stem ( ⁇ 10 nucleotides) extending beyond the Drosha cleavage site may be essential for efficient processing.
- Drosha is an RNase III endonuclease.
- Drosha may recognize terminal loops in the pri-miRNA and cleave approximately two helical turns into the stem to produce a 60-70 nucleotide precursor known
- the pre-miRNA may then be actively transported from the nucleus to the cytoplasm by Ran-GTP and the export receptor Ex-portin-5.
- the pre-miRNA may be recognized by Dicer, which is also an RNase III endonuclease. Dicer may recognize the double-stranded stem of the pre-miRNA. Dicer may also recognize the 5' phosphate and 3' overhang at the base of the stem loop. Dicer may cleave off the terminal loop two helical turns away from the base of the stem loop leaving an additional 5' phosphate and ⁇ 2 nucleotide 3' overhang.
- the resulting siRNA-like duplex which may comprise mismatches, comprises the mature miRNA and a similar-sized fragment known as the miRNA*.
- the miRNA and miRNA* may be derived from opposing arms of the pri-miRNA and pre-miRNA. MiRNA* sequences may be found in libraries of cloned miRNAs but typically at lower frequency than the miRNAs
- RISC RNA-induced silencing complex
- the miRNA* When the miRNA strand of the miRNA:miRNA* duplex is loaded into the RISC, the miRNA* may be removed and degraded.
- the strand of the miRNA:miRNA* duplex that is loaded into the RISC may be the strand whose 5' end is less tightly paired, hi cases where both ends of the miRNA:miRNA* have roughly equivalent 5' pairing, both miRNA and miRNA* may have gene silencing activity.
- the RISC may identify target nucleic acids based on high levels of complementarity between the miRNA and the mRNA, especially by nucleotides 2-7 of the miRNA. Only one case has been reported in animals where the interaction between the miRNA and its target was along the entire length of the miRNA. This was shown for mir-196 and Hox B 8 and it was further shown that mir-196 mediates the cleavage of the Hox B8 mRNA (Yekta et al 2004, Science 304-594). Otherwise, such interactions are known only in plants (Bartel & Bartel 2003, Plant Physiol 132-709).
- miRNAs may direct the RISC to downregulate gene expression by either of two mechanisms: mRNA cleavage or translational repression.
- the miRNA may specify cleavage of the mRNA if the mRNA has a certain degree of complementarity to the miRNA. When a miRNA guides cleavage, the cut may be between the nucleotides pairing to residues 10 and 11 of the miRNA. Alternatively, the miRNA may repress translation if the miRNA does not have the requisite degree of complementarity to the miRNA. Translational repression may be more prevalent in animals since animals may have a lower degree of complementarity between the miRNA and the binding site.
- any pair of miRNA and miRNA* may be due to variability in the enzymatic processing of Drosha and Dicer with respect to the site of cleavage. Variability at the 5' and 3' ends of miRNA and miRNA* may also be due to mismatches in the stem structures of the pri- miRNA and pre-miRNA. The mismatches of the stem strands may lead to a population of different hairpin structures. Variability in the stem structures may also lead to variability in the products of cleavage by Drosha and Dicer.
- nucleic acids are provided herein.
- the nucleic acids of the invention comprise the sequence of the provided nucleic acids or variants thereof.
- the variant may be a complement of the referenced nucleotide sequence.
- the variant may also be a nucleotide sequence that is substantially identical to the referenced nucleotide sequence or the complement thereof.
- the variant may also be a nucleotide sequence which hybridizes under stringent conditions to the referenced nucleotide sequence, complements thereof, or nucleotide sequences substantially identical thereto.
- the nucleic acid may have a length of from 10 to 250 nucleotides.
- the nucleic acid may have a length of at least 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 35, 40, 45, 50, 60, 70, 80, 90, 100, 125, 150, 175, 200 or 250 nucleotides.
- the nucleic acid may be synthesized or expressed in a cell (in vitro or in vivo) using a synthetic gene described herein.
- the nucleic acid may be synthesized as a single strand molecule and hybridized to a substantially complementary nucleic acid to form a duplex.
- the nucleic acid may be introduced to a cell, tissue or organ in a single- or double-stranded form or capable of being expressed by a synthetic gene using methods well known to those skilled in the art, including as described in U.S. Patent No. 6,506,559 which is incorporated by reference. 3a. Nucleic acid complexes
- the nucleic acid may further comprise one or more of the following: a peptide, a protein, a RNA-DNA hybrid, an antibody, an antibody fragment, a Fab fragment, and an aptamer.
- the nucleic acid may comprise a sequence of a pri-miRNA or a variant thereof.
- the pri-miRNA sequence may comprise from 45-30,000, 50-25,000, 100-20,000, 1,000-1,500 or 80-100 nucleotides.
- the sequence of the pri-miRNA may comprise a pre-miRNA, miRNA and miRNA*, as set forth herein, and variants thereof.
- the pri-miRNA may form a hairpin structure.
- the hairpin may comprise a first and a second nucleic acid sequence that are substantially complimentary.
- the first and second nucleic acid sequence may be from 37-50 nucleotides.
- the first and second nucleic acid sequence may be separated by a third sequence of from 8-12 nucleotides.
- the hairpin structure may have a free energy of less than -25 Kcal/mole, as calculated by the Vienna algorithm, with default parameters as described in Hofacker et al., Monatshefte f. Chemie 125: 167-188 (1994), the contents of which are incorporated herein.
- the hairpin may comprise a terminal loop of 4-20, 8-12 or 10 nucleotides.
- the pri-miRNA may comprise at least 19% adenosine nucleotides, at least 16% cytosine nucleotides, at least 23% thymine nucleotides and at least 19% guanine nucleotides. 3c. Pre-miRNA
- the nucleic acid may also comprise a sequence of a pre-miRNA or a variant thereof.
- the pre-miRNA sequence may comprise from 45-90, 60-80 or 60-70 nucleotides.
- the sequence of the pre-miRNA may comprise a miRNA and a miRNA* as set forth herein.
- the sequence of the pre-miRNA may also be that of a pri-miRNA excluding from 0-160 nucleotides from the 5' and 3' ends of the pri-miRNA. 3d. miRNA
- the nucleic acid may also comprise a sequence of a miRNA (including miRNA*) or a variant thereof.
- the miRNA sequence may comprise from 13-33, 18-24 or 21-23 nucleotides.
- the miRNA may also comprise a total of at least 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39 or 40 nucleotides.
- the sequence of the miRNA may be the first 13-33 nucleotides of the pre-miRNA.
- the sequence of the miRNA may also be the last 13-33 nucleotides of the pre-miRNA.
- the nucleic acid may also comprise a sequence of an anti-miRNA capable of blocking the activity of a miRNA or miRNA*, such as by binding to the pri-miRNA, pre- miRNA, miRNA or miRNA* (e.g. antisense or RNA silencing), or by binding to the target binding site.
- the anti-miRNA may comprise a total of 5-100 or 10-60 nucleotides.
- the anti-miRNA may also comprise a total of at least 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39 or 40 nucleotides.
- the sequence of the anti-miRNA may comprise (a) at least 5 nucleotides that are substantially identical or complimentary to the 5' of a miRNA and at least 5-12 nucleotides that are substantially complimentary to the flanking regions of the target site from the 5' end of the miRNA, or (b) at least 5-12 nucleotides that are substantially identical or complimentary to the 3' of a miRNA and at least 5 nucleotide that are substantially complimentary to the flanking region of the target site from the 3' end of the miRNA.
- the nucleic acid may also comprise a sequence of a target microRNA binding site or a variant thereof.
- the target site sequence may comprise a total of 5-100 or 10-60 nucleotides.
- the target site sequence may also comprise a total of at least 5, 6, 7, 8, 9, 10, 11, 12, 13, 14,
- a probe may comprise a nucleic acid.
- the probe may have a length of from 8 to 500, 10 to 100 or 20 to 60 nucleotides.
- the probe may also have a length of at least 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 35, 40, 45, 50, 60, 70, 80, 90, 100, 120, 140, 160, 180, 200, 220, 240, 260, 280 or 300 nucleotides.
- the probe may comprise a nucleic acid of 18-25 nucleotides.
- the sequence of the probes of the invention may comprise the sequence of SEQ ID NOS: 48-50 or variants thereof.
- a probe may be capable of binding to a target nucleic acid of complementary sequence through one or more types of chemical bonds, usually through complementary base pairing, usually through hydrogen bond formation. Probes may bind target sequences lacking complete complementarity with the probe sequence depending upon the stringency of the hybridization conditions.
- a probe may be single stranded or partially single and partially double stranded. The strandedness of the probe is dictated by the structure, composition, and properties of the target sequence. Probes may be directly labeled or indirectly labeled.
- the probe may further comprise a linker.
- the linker may be 10-60 nucleotides in length.
- the linker may be 20-27 nucleotides in length.
- the linker may be of sufficient length to allow the probe to be a total length of 45-60 nucleotides.
- the linker may not be capable of forming a stable secondary structure, or may not be capable of folding on itself, or may not be capable of folding on a non-linker portion of a nucleic acid contained in the probe.
- Target sequences of a cDNA may be generated by reverse transcription of the target KNA.
- Methods for generating cDNA may be reverse transcribing polyadenylated RNA or alternatively, RNA with a ligated adaptor sequence.
- the RNA may be ligated to an adaptor sequence prior to reverse transcription.
- a ligation reaction may be performed by T4 RNA ligase to ligate an adaptor sequence at the 3' end of the RNA.
- Reverse transcription (RT) reaction may then be performed using a primer comprising a sequence that is reverse complementary to the 3' end of the adaptor sequence.
- Polyadenylated RNA may be used in a reverse transcription (RT) reaction using a poly(T) primer (also referred to as oligo dT) comprising a 5' adaptor sequence.
- the poly(T) sequence may comprise 8, 9, 10, 11, 12, 13, or 14 consecutive thymines.
- the reverse transcript of the RNA may be amplified by real time PCR, using a specific forward primer comprising at least 15 nucleic acids complementary to the target nucleic acid and a 5' tail sequence; a reverse primer that is complementary to the 3' end of the adaptor sequence; and a probe that may be specifically complementary to the target nucleic acid.
- the probe may be partially complementary to the 5' end of the adaptor sequence.
- Methods of amplifying target nucleic acids are described herein.
- the amplification may be by a method comprising PCR.
- the first cycles of the PCR reaction may have an annealing temp of 56°C, 57°C, 58°C, 59°C, or 60°C.
- the first cycles may comprise 1-10 cycles.
- the remaining cycles of the PCR reaction may be 6O 0 C.
- the remaining cycles may comprise 2-40 cycles.
- the annealing temperature may cause the PCR to be more sensitive.
- the PCR may generate longer products that can serve as higher string
- the PCR reaction may comprise a forward primer.
- the forward primer may comprise 15, 16, 17, 18, 19, 20, or 21 nucleotides identical to the target nucleic acid.
- the 3' end of the forward primer may be sensitive to differences in sequence between a target nucleic acid and a sibling nucleic acid.
- the forward primer may also comprise a 5' overhanging tail.
- the 5' tail may increase the melting temperature of the forward primer.
- the sequence of the 5' tail may comprise a sequence that is non-identical to the genome of the animal from which the target nucleic acid is isolated.
- the sequence of the 5' tail may also be synthetic.
- the 5' tail may comprise 8, 9, 10, 11, 12, 13, 14, 15, or 16 nucleotides.
- the PCR reaction may comprise a reverse primer.
- the reverse primer may be complementary to a target nucleic acid.
- the reverse primer may also comprise a sequence complementary to an adaptor sequence.
- the sequence complementary to an adaptor sequence may comprise 12-24 nucleotides.
- kits may comprise a nucleic acid described herein together with any or all of the following: assay reagents, buffers, probes and/or primers, and sterile saline or another pharmaceutically acceptable emulsion and suspension base.
- the kits may include instructional materials containing directions (e.g., protocols) for the practice of the methods described herein.
- the kit may be used for the amplification, detection, identification or quantification of a target nucleic acid sequence.
- the kit may comprise a poly(T) primer, a forward primer, a reverse primer, and a probe. Any of the compositions described herein may be comprised in a kit.
- reagents for isolating miRNA, labeling miRNA, and/or evaluating a miRNA population using an array are included in a kit.
- the kit may further include reagents for creating or synthesizing miRNA probes.
- the kits will thus comprise, in suitable container means, an enzyme for labeling the miRNA by incorporating labeled nucleotide or unlabeled nucleotides that are subsequently labeled.
- kits of the invention may include components for making a nucleic acid array comprising miRNA, and thus, may include, for example, a solid support.
- Example 1 miRNA detection by the General Probe Detection assay, SYBRGreen, and Varkonyi-Gasic (stem-loop) techniques hsa-miR-296-3 ⁇ (SEQ. ID NO: 11) was detected by the General (antisense) Probe
- RNA isolation and cDNA preparation 1.1.2.
- RNA isolation and cDNA preparation 1.1.2.
- Real-time PCR " RTPCR) reactions
- RNA 1 ⁇ g was polyadenylated in a 10 ⁇ l reaction containing 2.5 niM MnCl 2 , PNK buffer (New England Biolabs, Ipswich, MA), 4mM ATP (Amersham Pharmacia, Uppsala, Sweden) and 1.5 units of poly A polymerase (Takara,
- RTl primer SEQ. ID NO: 26
- sequence of this RTl primer is composed of a 3' rapid amplification of complementary DNA ends tail, and the UPL#61 sequence.
- the reaction was heated to 85°c for 2 min in order to disrupt any existing RNA secondary structures, followed by temperature gradient in which the temperature decreased gradually from 75°c to 25°c.
- Each sample was then combined with 12 ⁇ l of RT mix, to a final volume of 20 ⁇ l, containing 0.125mM of each dNTP (Amersham, Uppsala, Sweden), 0.255M trealose, 5x RT buffer, 1OmM DTT and 200 units of Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA).
- minus-RT control For each sample a control which did not contain reverse transcriptase was prepared (further referred to as minus-RT control).
- the reaction was heated for 5 min at 37°c and 5 min at 45°c for 5 cycles.
- the real-time PCR reactions of the General Probe Detection Assay were performed in lO ⁇ l volume reaction which included 5 ⁇ l ABsoluteTM qPCR mix (ABgene, IL, USA), 0.5 ⁇ M miR-specific forward primer (SEQ. ID NO: 12, for hsa-miR-296-3 ⁇ , and SEQ. ID NO: 8, for hsa-miR-181a), 0.5 ⁇ M reverse primer (SEQ. ID NO: 28), and 0.05 ⁇ M UPL#61 (SEQ. ID NO: 50).
- the reactions were incubated at 95°c for 15 min followed by 45 cycles of 95°c for 15s, 60°c for 20s and 60°c for 20s, and then cooled to 40°c.
- PoIyA based analysis of miRNAs was done using SYBR Green as described above except for the use of a different RT primer, RT2 (SEQ. ID NO: 27), which lacks the UPL#61 binding sequence.
- the real-time PCR reactions of the SYBR Green analysis was performed in a 10 ⁇ l volume reaction which included 2X Power SYBR ® Green PCR master mix (Applied Biosystems, Foster City, CA), 0.5 ⁇ M forward hsa-miR-296-3p specific primer (SEQ. ID NO: 12) and 0.5 ⁇ M reverse primer (SEQ. ID NO: 28).
- the reactions were incubated at 95°c for 10 min followed by 45 cycles of 95°c for 10s and 60°c for 1 min. For melting curve analysis the reactions were heated to 95°c for 5s and 60°c for 15s. The reactions were then cooled to 40 0 C.
- Varkonyi-Gasic method for miRNA detection is done in one tube and does not involve cDNA preparation.
- the real-time PCR reactions were conducted as follows: Stem-loop qPCR amplification of miR-181a (SEQ. TD NO: 7) was performed using RNA UltrasenseTM One- Step RT-PCR kit (hwitrogen, Carlsbad, CA) as described (Varkonyi-Gasic et al, 2007). The RT, the forward, and the reverse primers sequences are provided in table 1. UPL#61 (SEQ. ID NO: 50) was added at a final concentration of 0.05 ⁇ M. Reaction was carried out as follows: 50°c for 15 min, 95°c for 2 min, followed by 45 cycles of 95°c for 15s and 60°c for 30s.
- Example 1.2 Comparison of the detection hsa-miR-296-3p (SEQ. ID NO: 11) by the General Probe Detection assay with its detection by SYBR Green hsa-miR-296-3p (SEQ. ID NO: 11) was amplified using both of the General Probe
- Example 1.3 Comparison of the detection of hsa-miR-181a (SEQ. ID NO: 7) by the General Probe Detection assay with its detection by the Varkonyi-Gasic (stem-loop) method
- a miRNA family is typically characterized by single to a few nucleotides differences within the 3'-end of its miRNAs members.
- the ability of the General (antisense) Probe Detection assay to discriminate miRNAs within a family was tested on the let-7 family.
- Two cDNA libraries were prepared from synthetic miRNA sequences of let-7c and let-7 d, and were then amplified using the sequences of hsa-let-7a to -7e as forward primers. Relative detection specificity the General (antisense) Probe Detection assay for each forward primer was calculated as a percent of the perfect match of the Cp values of target and off-target assays (100x2 " ⁇ Cp ) when using 250pg of cDNA prepared from synthetic RNA.
- let-7c cDNA As depicted in table 2, very low signals of non-specific amplification were observed for let-7c cDNA when it was tested with the forward primers let-7a (SEQ. ID NO: 14), let- 7b (SEQ. ID NO: 16), let-7d (SEQ. ID NO: 20), and let-7e (SEQ. ID NO: 22) (0.3%, 0.8%, 0%, and 0%, respectively).
- the let-7d cDNA gave similar results except for let-7a (SEQ. ID NO: 14), where the assay could detect 32 % of the target sequence, probably as a result of the wobble at the 5'end of the RT primer.
- the sensitivity of the General Probe Detection assay and its ability to detect specific viral miRNAs in the presence of other host miRNAs were determined in a competition assay.
- One ⁇ g of total RNA isolated from B95-8 cells (a B cell line infected by EBV) and 1 ⁇ g of total RNA isolated from UKF-NB4 cells were reverse transcribed separately.
- the cDNA library of the B95-8 cells was serially diluted by two folds, from 250 to 0.25 pg, with the cDNA library of the UKF-NB4 cells.
- the diluted B-95 library was then used in the General (antisense) Probe Detection assay to detect EBV-miR-BARTl (SEQ.
- EBV-miR-BARTl SEQ. ID NO: 23
- EBV-BARTl SEQ. ID NO: 23
- Fig. 4A originated from B-95-8 total RNA (in total of 0.25 ng cDNA).
- Fig. 4B no fluorescence was detected in the -RT control or in the no-template negative controls (Fig. 4B).
- the General (antisense) Probe Detection assay showed excellent linearity over five orders of magnitude ranging from 0.25 to 2500 pg. As indicated in Figure 5A, it was possible to detect hsa-miR-30a (SEQ. ID NO: 1), hsa-miR-103 (SEQ. ID NO: 3), hsa-miR- 107 (SEQ. ID NO: 5), hsa-miR-181a (SEQ. ID NO: 7), and hsa-miR-210 (SEQ. ID NO: 9) in as little as 0.25 ⁇ g of total RNA.
- the EBV-miRNAs BART3 SEQ.
- BHRF1-2 SEQ. ID NO: 35
- BART3* SEQ. ID NO: 34
- BHRF1-2* SEQ. ID NO: 36
- the General (antisense) Probe Detection assay could detect miRNAs, which are not highly abundant, from a single cell.
- RNA extracted from Peripheral blood mononuclear cells isolated from 10 ml of whole blood with FicoU-Hypaque solution (GE Healthcare Biosciences, Uppsala, Sweden). The details of sequences are presented in table 4.
- a specific primer for U6 (SEQ. ID NO: 25) was used for normalization.
- the PBMCs were collected from three individuals: one of whom had an acute EBV infection [infectious mononucleosis (IM, with the following serological markers: positive for EBV-VCA IgG and IgM but negative for EBV-EBNA antibodies)], and two healthy carriers of EBV (positive for IgG antibodies to EBV-EBNA and EBV-VCA and negative for IgM VCA, indicating past and latent EBV infection). Since the expected number of B- cells that harbor EBV in PBMCs of healthy donors is estimated to be one out of 10 6 cells, and RNA was extracted from 10 7 PBMCs, the qPCR cycling of the General Probe Detection Assay was extended to 75 cycles instead of the usual 40 cycles that were used otherwise.
- IM infectious mononucleosis
- the assay showed a very high sensitivity; it was able to detect EBV-miRNAs in PBMCs of both the patient and the healthy carriers of EBV.
- a differential expression of various EBV-miRNAs was observed during the acute IM vs. the latent infection (healthy donors): BART2 (SEQ. ID NO: 32), BART4 (SEQ. ID NO: 37), and BART6 (SEQ. ID NO: 38) were clearly expressed in healthy carriers only, while BART7 (SEQ. ID NO: 39) was expressed only in the acute M. Regardless of the 75 cycles used in the qPCR, the "no-template-control" remained negative.
- Chlamydomonas reinhardtii algae, UTEX 90 strain (University of Texas algae collection, Austin TX) were maintained in TAP medium in an Adaptis growth chamber
- the extracted material was treated by DNase (Ambion), extracted by phenol and participated by ETOH.
- Cre-mirR1142 (SEQ. ID NO: 51) was detected by both of the SYBR Green method and the General (antisense) Probe Detection Assay.
- the General probe used in this example was used in this example.
- SEQ. ID NO: 48 is an MGB probe which is partially complementary to the adaptor sequence region of the sense strand of the amplicon and further partially complementary to the poly (T) region of the sense strand of the amplicon.
- RNA was incubated in the presence of a poly A polymerase enzyme (Takara, Otsu Japan), MnCl 2 , and ATP for 1 h at 37°C. Then, using an oligo dT tail harboring a consensus sequence, reverse transcription was performed on total RNA
- RNA using Superscript II RT (Invitrogen, Carlsbad CA).
- the cDNA was amplified by real time PCR; this reaction contained a microRNA-specific forward primer and a universal reverse primer complementary to the consensus 3' sequence of the oligo dT tail.
- the sequences used in the PCR procedure are detailed in table 5 below.
- V represents A, G or C, and N represents any nucleotide.
- Example 4 miR detection: with a specific probe, by the General (antisense) Probe Detection Assay and by the General (antisense) Probe Detection Assay with a forward primer harboring an elongated tail sequence
- RNA material Five samples of stomach tissue were obtained; FFPE samples were incubated repeatedly in xylene at 57 0 C to remove excess paraffin, followed by washing in ethanol. Proteins were digested by proteinase K solution at 45 0 C for a few hours. The RNA was extracted with acid phenol: chloroform, followed by ethanol precipitation and DNAse digestion. cDNA was prepared from the RNA. Each cDNA was diluted to 5 ng/ ⁇ l and than to
- the target nucleic acid, hsa-miR-451 (most abundant variant: SEQ. ID NO: 54) was detected in the cDNA by the three following methods:
- the same reverse primer (SEQ. ID NO: 28) was used in all of the qPCR reactions.
- the General Probe Assay (sense and antisense) allows for detection of multiple variants of the targeted miR, as opposed to the assay with the specific probe, which detects only a specific variant.
- the sequences used in the detection are detailed in table 6.
- the qPCR reaction included 42 cycles.
- Table 7.2 detection of hsa-miR-451 (SEQ. ID NO: 54) (in Cts, average of triplicates) with a General antisense probe
- the average Ct, in both dilutions, is lower for the detection with the General probe than for the detection with the specific probe.
- This result implies that the hybridization of the forward primer used in the General (antisense) Probe Detection Assay to the target nucleic acid is improved as compared to the hybridization of the forward primer used in the detection with the specific probe, thereby enhancing the effectiveness of the detection.
- the average Ct (in both dilutions) is lowest for the detection with the General probe using a forward primer with an elongated tail. The elongation of the tail raises the T M of the forward primer to 6O 0 C at least.
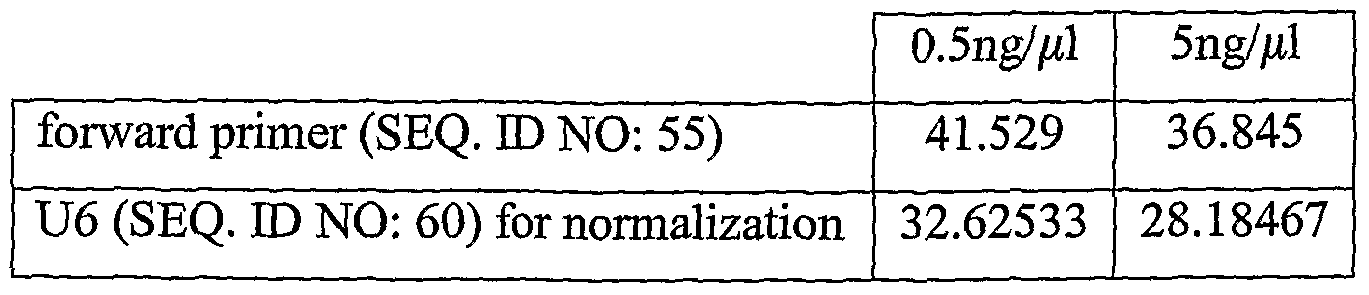
- Synthetic hsa-miR-451 (SEQ. ID NO: 59) was used to prepare 2.5nM cDNA.
- Synthetic hsa-miR-451 (SEQ. ID NO: 59) was detected with a specific probe (SEQ. ID NO: 56) and a forward primer (SEQ. ID NO: 55), and also with the General antisense MGB Probe (SEQ. ID NO: 48, which is partially complementary to the adaptor sequence region of the sense strand and to the poly(T) region of the sense strand of the amplicon) and a forward primer with an elongated tail (SEQ. ID NO: 58).
- the same reverse primer (SEQ. ID NO: 28) was used in both qPCR reactions.
- the average Ct, in all of the dilutions, is lower for the detection with the General antisense Probe than for the detection with the specific probe.
- This finding is in accordance with the results obtained in the detection of hsa-miR-451 (SEQ. ID NO: 54) in tissue samples as described in Example 4.1 above; this negates the possibility that the low Ct's of the General (antisense) Probe detection Assay are the result of non-specific detection of nucleic acids, since no other nucleic acids besides the synthetic hsa-miR-451 (SEQ. ID NO: 59) were in the tested samples.
- Example 5 General Probe Detection Assay: detection with a General sense Probe
- the probe of the General Probe Detection Assay is an MGB probe which is complementary to the adaptor sequence region and to the poly(T) region of the antisense strand of the amplicon. This probe is designated General sense Probe, and it may be reverse complement to the General antisense probe described in examples 3 and 4 above.
- RNA from fresh liver samples and from brain samples was obtained from Ambion.
- cDNA was prepared from the RNA, and serially diluted in the cDNA of the other tissue to a final concentration of 0.5 ng/ ⁇ g liver-brain cDNA mixture.
- the General (sense) Probe Detection Assay was then used to detect each of the liver-specific hsa-miR-122 (SEQ. ID NO: 61) and the brain-specific hsa-miR-124 (SEQ. ID NO: 62) in the mixtures.
- Table 11.1 Detection of liver-specific hsa-miR-122 (SEQ. ID NO: 61) spiked in cDNA of Brain tissues
- Table 11.2 Detection of brain-specific hsa-miR-124 (SEQ. ID NO: 62) spiked in cDNA of Liver tissues
- This sensitivity of the General Probe Detection Assay may be highly advantageous, for example, in the detection of minute concentrations of tumor tissue metastases to other tissue or organ.
- Table 13 Cross reactivity (in Cts) between members of the hsa-let-7 family, as detected with the sense probe of the General Probe Detection Assay
- General sense MGB Probe to detect each of the specific synthetic sequences in a mixture with other family members. This is essential for detection of specific biomarker-miRs in tissues comprising multiple miRs.
- RNA extraction 100 ml serum was incubated at 56 0 C for 1 h with 0.65 mg/ml Proteinase K (Sigma P2308). Two synthetic RNAs (IDT) were spiked-in as controls before acid phenolxhloroform extraction and then RNA was ETOH precipitated overnight at - 20 0 C.
- IDT Two synthetic RNAs
Landscapes
- Chemical & Material Sciences (AREA)
- Organic Chemistry (AREA)
- Life Sciences & Earth Sciences (AREA)
- Zoology (AREA)
- Wood Science & Technology (AREA)
- Proteomics, Peptides & Aminoacids (AREA)
- Health & Medical Sciences (AREA)
- Engineering & Computer Science (AREA)
- Microbiology (AREA)
- Immunology (AREA)
- Physics & Mathematics (AREA)
- Molecular Biology (AREA)
- Biotechnology (AREA)
- Biophysics (AREA)
- Analytical Chemistry (AREA)
- Biochemistry (AREA)
- Bioinformatics & Cheminformatics (AREA)
- General Engineering & Computer Science (AREA)
- General Health & Medical Sciences (AREA)
- Genetics & Genomics (AREA)
- Measuring Or Testing Involving Enzymes Or Micro-Organisms (AREA)
Abstract
The present invention provides a method to detect and quantify RNAs with high sensitivity and specificity. The method makes use of polyadenylation, primers, and general probes for the detection of RNA sequences including mRNAs and short RNA sequences such as miRNA.
Description
METHOD FOR DETECTION OF NUCLEIC ACID SEQUENCES
CROSS REFERENCE TO RELATED APPLICATIONS
The present application claims priority under 35 U.S.C. § 119(e) to U.S. Provisional Application No. 61/158,776, filed March 10, 2009, which is herein incorporated by reference in its entirety.
FIELD OF THE INVENTION
The invention relates in general to a method for detecting RNA sequences, particularly but not limited to microRNA and other short RNA sequences.
BACKGROUND OF THE INVENTION
MicroRNAs (miRNAs) are an abundant class of short (~22nt) non-coding single- stranded RNAs. They typically bind to complementary sequences at the 3 '-untranslated region (UTR) of mRNAs and thereby exert postranscriptional regulation on gene expression and protein translation. There is increasing evidence suggesting that miRNAs play critical roles in many key biological processes, such as cell growth, tissue differentiation, cell proliferation, embryonic development, apoptosis, viral infection, cancer and more. Frequently miRNAs expression is tissue specific so much so that they were suggested to serve as biomarkers and to accurately identify cancers of unknown tissue origin.
In order to further investigate the role of miRNAs in specific tissues and cells and to use them as tools for research and clinical diagnosis, efficient and reliable detection methods are required. Standard technologies such as cloning, northern hybridization and microarray analysis are time consuming, and are not sensitive enough to detect less abundant miRNA.
Real time PCR (RTPCR) is a simple and highly sensitive technique, extensively used for gene expression and quantification. However, the short length of miRNAs present a challenge for RTPCR test and various solutions were offered to solve this problem. To date two approaches have been most commonly used to detect miRNAs by RTPCR. In the first, stem-loop primers are used to reverse transcribe (RT) the miRNA, and the RT product is quantified in a TaqMan™ PCR (qRTPCR) assay) (Chen, C. Nucleic Acids Research, Vol. 33, No. 20, 2005). Although this method is highly sensitive and accurate, it requires an individual miRNA-specific fluorescent probe, making it highly expensive. As a result, this
technique may not be applicable for screening of large number of miRNAs for most research laboratories nor can it serve as a future miRNA-based diagnosis tool.
Varkonyi-Gasic et al {Plant Methods 3:12, 2007) have detected and quantified miRNAs by stem loop reverse primer and qRTPCR using a generic and small universal probe. The Varkonyi-Gasic method is based on the method first described by Chen et al., with replacement of the specific probe by a generic probe. This one step method also requires a unique stem-loop-specific reverse transcription primer for each miRNA in addition to the miRNA specific forward primer. This generic primer is 50nts long, and thus requires a special synthesis scale. Furthermore, amplification is limited to 35 cycles due to non-specific amplification in higher cycles.
The second technique relays on the use of SYBR Green for detection of miRNAs. Since SYBR Green intercalates into double-stranded DNA in a non-specific manner, it commonly detects non-specific PCR products and primer-dimers, leading to false-positive results. Due to the small size of these products, in most cases melting curve analysis and gel electrophoresis are unable to distinguish between the specific and non-specific PCR products. For these reasons the PCR-product can not be quantified directly.
There is an unmet need for a reliable inexpensive, sensitive and specific method for the detection of RNA, particularly mRNA and miRNA.
SUMMARY OF THE INVENTION
The present invention provides a method of detecting a target RNA nucleic acid sequence in a biological sample, the method comprising: providing the biological sample comprising the target RNA nucleic acid sequence, annealing the target RNA nucleic acid sequence with a poly(T) primer comprising a 5' universal adaptor sequence, generating a reverse transcript of the polyadenylated RNA, with the poly(T) primer, and amplifying the reverse transcript product by a polymerase chain reaction (PCR) comprising a specific forward primer, a universal reverse primer and a general probe, to generate an amplicon; wherein the forward primer is at least partially identical to the target RNA nucleic acid sequence and the universal reverse primer is at least partially identical to a 5' region of the adaptor sequence of the poly (T) primer.
According to some embodiments the target RNA nucleic acid sequence is a short nucleic acid, and it is extended at the 3' end by polyadenylation, prior to its annealing with the poly(T) primer.
The present invention further provides a kit for the detection of a target RNA nucleic acid sequence in a biological sample, the kit comprising a poly(T) primer comprising a 51 universal adaptor sequence, a specific forward primer, a universal reverse primer and a general probe, wherein the forward primer is at least partially identical to the target RNA nucleic acid sequence, and the universal reverse primer is at least partially identical to a 5' region of the adaptor sequence of the poly (T) primer.
According to some embodiments the general probe is partially complementary to the adaptor sequence region of the sense strand of the amplicon. According to some embodiments the general probe is partially further partially complementary to the poly(T) region of the sense strand of the amplicon. According to additional embodiments the general probe is complementary to the adaptor sequence region of the antisense strand of the amplicon. According to some embodiments the general probe is further partially complementary to the poly(T) region of the antisense strand of the amplicon. According to some embodiments the general probe comprises a sequence selected from the, group consisting of SEQ. ID NOS: 48-50. According to some embodiments the general probe which is partially complementary to the adaptor sequence region of the sense strand of the amplicon comprises a sequence selected from the group consisting of SEQ. H) NOS: 48 and 50. According to some embodiments the general probe which is further partially complementary to the poly(T) region of the sense strand of the amplicon comprises SEQ. ID. NO: 48. According to other embodiments the general probe which is complementary to the adaptor sequence region of the antisense strand of the amplicon, and further partially complementary to the poly(T) region of the antisense strand of the amplicon comprises SEQ. ID. NO: 49.
According to some embodiments the forward primer comprises a sequence selected from the group consisting of SEQ. ID NOS. 2, 4, 6, 8, 12, 14, 16, 18, 20, 22, 24, 40-47, 52, 55, 57, 58, 63, 64, 69-87, 89 and 91. According to some embodiments the reverse primer comprises the sequence of SEQ. ID NO. 28. According to some embodiments the method of the invention comprises SEQ. ID NO. 60 as a normalizer.
According to some embodiments the target RNA nucleic acid sequence is selected from the group consisting of: mRNA, microRNA, siRNA, PiwiRNA, and a combination thereof. According to some embodiments the target RNA nucleic acid sequence is a short RNA nucleic acid sequence selected from the group consisting of: microRNA, siRNA, PiwiRNA, and a combination thereof. According to some embodiments the target RNA nucleic acid sequence is a short RNA nucleic acid sequence 17-25 nt in length.
According to some embodiments the general probe comprises a fluorescent reporter group and a quencher. The general probe may comprise a 5' fluorescent reporter group and 3' a quencher. According to other embodiments the general probe may comprise a 3' fluorescent reporter group and a 5' quencher. According to some embodiments the general probe is a TaqMan probe. According to some embodiments the general probe further comprises a minor groove binder (MGB). According to other embodiments the general probe further comprises locked nucleic acids (LNA).
According to some embodiments the biological sample is selected from the group consisting of bodily fluid, a cell line, a tissue sample, a biopsy sample, a needle biopsy sample, a surgically removed sample, and a sample obtained by tissue-sampling procedures. According to some embodiments the bodily fluid is serum. According to some embodiments the tissue is a fresh, frozen, fixed, wax-embedded or formalin-fixed paraffin- embedded (FFPE) tissue. According to other embodiments the biological sample is derived from a plant. These and other embodiments of the present invention will become apparent in conjunction with the figures, description and claims that follow.
BRIEF DESCRIPTION OF THE DRAWINGS
Figure IA Schematic description of the General (antisense) Probe Detection Assay of the invention.
Step 1 (top vertical arrow): polyadenylation of total RNA to generate poly(A) miRNA.
Step 2 (middle vertical arrow): reverse transcription of poly(A) miRNA using a poly(T) primer that contains a universal tail (adaptor) and a probe binding region.
Step 3 (bottom vertical arrow): Real Time PCR amplification using a miRNA-specific forward primer, a reverse primer complementary to the universal tail and a general antisense probe.
Figure IB Schematic description of the Varkonyi-Gasic (stem loop) assay.
Step 1 (top vertical arrow): stem-loop RT primers bind to the 3' portion of the miRNA and are reverse transcribed; Step 2 (bottom vertical arrow): PCR amplification using miRNA-specific forward primer, specific reverse primer and a TaqMan probe.
Figure 2. Figures 2A-2D present amplification curves, in which the y-axis represents fluorescence (483-533) and the x-axis represents the number of cycles. The amounts of cDNA/RNA used for each reaction were as follows: (1) 2.5 ng; (2) 250 pg; (3) 25 pg; (4)
2.5 pg; (5) 0.25 pg; (-RT, no reverse transcriptase negative control) 2.5 ng RNA; (NTC) no- template negative control. One experiment out of three is shown.
Fig. 2A Detection of hsa-rαiR-296-3p (SEQ. ID NO: 11) by qPCR (quantitative polymerase chain reaction) using SYBR Green (Slope =~3.8517; E = 0.8181)
Fig. 2B Detection of hsa-miR-296-3p (SEQ. ID NO: 11) by qPCR using the General (antisense) Probe Detection assay with universal probe library (UPL#61, SEQ. ID NO: 50) (Slope =-3.9633; E = 0.7878)
Fig. 2C Detection of hsa-miR-181a (SEQ. ID NO: 7) by qPCR using the General (antisense) Probe Detection assay with universal probe library (UPL #61, SEQ. ID NO: 50) (Slope =-3.5617; E = 0.9088).
Fig. 2D Stem-loop qRT-PCR-UPL amplification of hsa-miR-181a (SEQ. ID NO: 7), using RNA as a template (Slope =-3.1849; E = 1.0605).
Figure 3. Analysis of qPCR amplification on 2.5% agarose gel Fig. 3A SYBR-Green qPCR amplification of hsa-miR-296-3p (SEQ. ID NO: 11). The arrow indicates the location of the band of the mature miR.
Fig. 3B Varkonyi-Gasic stem-loop qPCR amplification of hsa-miR-181a (SEQ. ID NO: 7).
The arrow indicates the location of the band of the mature miR.
Fig. 3C General (antisense) Probe amplification: left- hsa-miR-181a (SEQ. ID NO: 7); right- hsa-miR-296-3p (SEQ. ID NO: 11). The arrow indicates the location of a band of
~70 nucleotides.
Figure 4. Detection of EBV-mir-BARTl (SEQ. ID NO: 23) in cDNA of UKF-NB4 by the
General (antisense) Probe Detection assay, in the presence of other miRs
Fig. 4A Correlation of B95-8 cDNA input to the AO-Cp value of the reaction (mean of three experiments). The x-axis represents B95-8 RNA (pg/reaction) and the y-axis AQ-Cp.
Fig. 4B Amplification curves of EBV-miRNA BARTl (SEQ. ID NO: 23) (one representative of three experiments, Slope =-3.3885; E = 0.973). The x-axis represents the number of cycles, and the y-axis - fluorescence (483-533). In each of the General Probe Detection Assay reactions a total of 250 pg of cDNA was amplified. B95-8 cDNA was diluted two folds from 250 pg (no UKF-cDNA) to 0.244 pg, in UKF-NB4-CDNA. Zero is UKF-cDNA only (no B95-8 cDNA). Amplification curves of RNA samples that were not reverse-transcribed, of B95-8, and of UKF-NB4 are shown as well (-RTB95 and -RT UKF-NB4). NTC is no-template negative control.
Figure 5. Validating detection of miRNAs using the General (antisense) Probe Detection assay
Fig. 5A Detection of various hsa-miRNAs using the General (antisense) Probe Detection Assay. Correlation of cDNA input (x-axis, cDNA libraries prepared from UKF-NB4 total RNA, pg/reaction) to the 40-Cp values (y-axis) for five host miRNAs. hsa-miR-30a (SEQ. ID NO: l) - x symbols; hsa-miR-103 (SEQ. ID NO: 3) - black square symbols; hsa-miR- 107 (SEQ. ID NO: 5) - white square symbols; hsa-miR-181a (SEQ. ID NO: 7) - triangles; hsa-miR-210 (SEQ. ID NO: 9) - circles.
Fig. 5B Correlation between B95-8 cDNA (pg/reaction) input (x-axis), and AQ-Cp values (y- axis) of two EBV-miRNAs [BART-3 (SEQ. ID NO: 33) and BHRF-1-2 (SEQ. ID NO: 35), black diamonds and squares, respectively] and their matching miRNAs* [BART-3* (SEQ. ID NO: 34) and BHRF-1-2* (SEQ. ID NO: 36), open black diamonds and squares, respectively], using cDNA libraries prepared from lμg of total RNA extracted from B95-8 cells. Figure 6. Expression of pattern of various EBV miRNAs in PBMCs in vivo. The x-axis depicts the EBV-miRNAs tested and the y-axis depicts the 75-Q? value for each sample. The General Probe Detection Assay for EBV-miRNAs was performed on RNAs extracted from ~107 PBMC/sample of two healthy carriers (white and grey bars) and an infectious mononucleosis patient (black bar); the no-template-controls for each of the EBV-miRs was negative.
Figure 7. Analysis of qPCR amplification on 2,5% agarose gel. The left arrow marks the lane with the cDNA including cre-miR1142 (SEQ. ID NO: 51); the right arrow marks the lane with water as a control.
Fig. 7 A qRT-PCR of products of the General Probe Detection assay Fig. 7B qRT-PCR of products of the SYBR green method
Figure 8. The sensitivity of the General (sense) Probe Detection Assay: a competition assay demonstrating the assays ability to detect specific miRs in the presence of others. Fig 8A Detection of liver-specific hsa-miR-122 (SEQ. ID NO: 61) in mixture of cDNA of total RNA from Liver and Brain tissues. The y-axis depicts the PCR signal normalized by subtracting it from 55 (55-Ct); the x-axis depicts the percentage of liver cDNA in the mixture of cDNA of total RNA from Liver and Brain tissues.
Fig 8B Detection of brain-specific hsa-miR-124 (SEQ. ID NO: 62) in mixture of cDNA of total RNA from Liver and Brain tissues.
The y-axis depicts the PCR signal normalized by subtracting it from 55 (55-Ct); the x-axis depicts the percentage of brain cDNA in the mixture of cDNA of total RNA from Liver and Brain tissues.
DETAILED DESCRIPTION OF THE INVENTION
The present invention provides a sensitive, specific and accurate method for the detection of RNA, including mRNAs and short RNAs such as miRNAs. In this method, designated General Probe Detection Assay, the RT and the reverse primers, as well as the probe (which may be sense or antisense), are generic, and the only specific component is the forward primer which determines the amplification of a specific RNA. The General Probe Detection assay is able to detect various viral, plant and human miRNAs in several cell lines with a very high sensitivity, and can also discriminate miRNAs that differ by as little as a single nucleotide, exhibiting high specificity. Furthermore, preparation of cDNA library and qRT-PCR by this method takes less than one day. This approach enables a versatile and cost effective detection of RNAs. Thus, this protocol can be applied to sensitive, robust, and rapid detection of miRNAs from various origins.
Before the present compositions and methods are disclosed and described, it is to be understood that the terminology used herein is for the purpose of describing particular embodiments only and is not intended to be limiting. It must be noted that, as used in the specification and the appended claims, the singular forms "a," "an" and "the" include plural referents unless the context clearly dictates otherwise.
For the recitation of numeric ranges herein, each intervening number there between with the same degree of precision is explicitly contemplated. For example, for the range of 6-9, the numbers 7 and 8 are contemplated in addition to 6 and 9, and for the range 6.0-7.0, the number 6.0, 6.1, 6.2, 6.3, 6.4, 6.5, 6.6, 6.7, 6.8, 6.9 and 7.0 are explicitly contemplated.
1. Definitions about
As used herein, the term "about" refers to +/-10%. adaptor
As used herein, adaptors are oligonucleotides that are continuous to a cDNA of a target nucleic acid, such that PCR primers may anneal (entirely or partially) thereto for
amplification of the target nucleic acid. Adaptor sequences may also be referred to as tail sequences. amplicon
As used herein, amplicons are fragments of DNA formed as the products of natural or artificial amplification. antisense
The term "antisense," as used herein, refers to nucleotide sequences which are reverse complementary to a specific DNA or RNA sequence. The term "antisense strand" is used in reference to a nucleic acid strand that is complementary to the "sense" strand. Antisense molecules may be produced by any method, including synthesis by ligating the gene(s) of interest in a reverse orientation to a viral promoter which permits the synthesis of a complementary strand. Once introduced into a cell, this transcribed strand combines with natural sequences produced by the cell to form duplexes. These duplexes then block either the further transcription or translation. In this manner, mutant phenotypes may be generated. antisense probe
The term "antisense probe" as used herein, refers to a probe which anneals to the cDNA polyadenylated strand (+ strand) of the amplicon comprising the sequence to be detected by the probe. The sequence of the antisense probe of the invention may comprise the sequences of SEQ. ID NOS: 48 and 50. attached
"Attached" or "immobilized" as used herein refer to a probe and a solid support and may mean that the binding between the probe and the solid support is sufficient to be stable under conditions of binding, washing, analysis, and removal. The binding may be covalent or non-covalent. Covalent bonds may be formed directly between the probe and the solid support or may be formed by a cross linker or by inclusion of a specific reactive group on either the solid support or the probe, or both. Non-covalent binding may be one or more of electrostatic, hydrophilic, and hydrophobic interactions. Included in non-covalent binding is the covalent attachment of a molecule, such as streptavidin, to the support and the non- covalent binding of a biotinylated probe to the streptavidin. Immobilization may also involve a combination of covalent and non-covalent interactions.
biological sample
"Biological sample" as used herein means a sample of biological tissue or fluid that comprises nucleic acids. Samples may originate from viruses, plants or animals including humans. Such samples include, but are not limited to, tissue or fluid isolated from subjects.
Biological samples may also include sections of tissues such as biopsy and autopsy samples, formalin-fixed paraffin-embedded (FFPE) samples, frozen sections taken for histological purposes, blood, plasma, serum, sputum, stool, tears, mucus, hair, and skin. Biological samples also include explants and primary and/or transformed cell cultures derived from animal or patient tissues .
Biological samples may also be blood, a blood fraction, urine, effusions, ascitic fluid, saliva, cerebrospinal fluid, cervical secretions, vaginal secretions, endometrial secretions, gastrointestinal secretions, bronchial secretions, sputum, cell line, tissue sample, cellular content of fine needle aspiration (FNA) or secretions from the breast. A biological sample may be provided by removing a sample of cells from a subject, but can also be accomplished by using previously isolated cells (e.g., isolated by another person, at another time, and/or for another purpose), or by performing the methods described herein in vivo. Archival tissues, such as those having treatment or outcome history, may also be used. complement "Complement" or "complementary" as used herein means Watson-Crick (e.g., A-
TAJ and C-G) or Hoogsteen base pairing between nucleotides or nucleotide analogs of nucleic acid molecules. A full complement or fully complementary may mean 100% complementary base pairing between nucleotides or nucleotide analogs of nucleic acid molecules. Partial complementary may mean less than 100% complementarity, for example 80% complementarity. In some embodiments, the complementary sequence has a reverse orientation (5 '-3').
Ct
"Ct" as used herein refers to Cycle Threshold of qRT-PCR, which is the fractional cycle number at which the fluorescence crosses the threshold. Cp is the crossing point. detection
"Detection" means detecting the presence of a component in a sample. Detection also means detecting the absence of a component. Detection also means measuring the level of a component, either quantitatively or qualitatively.
differential expression
"Differential expression" means qualitative or quantitative differences in the temporal and/or cellular gene expression patterns within and among cells and tissue. Thus, a differentially expressed gene may qualitatively have its expression altered, including an activation or inactivation, in, e.g., normal versus disease tissue. Genes may be turned on or turned off in a particular state, relative to another state thus permitting comparison of two or more states. A qualitatively regulated gene may exhibit an expression pattern within a state or cell type which may be detectable by standard techniques. Some genes may be expressed in one state or cell type, but not in both. Alternatively, the difference in expression may be quantitative, e.g., in that expression is modulated, either up-regulated- resulting in an increased amount of transcript, or down-regulated- resulting in a decreased amount of transcript. The degree to which expression differs need only be large enough to quantify via standard characterization techniques such as expression arrays, quantitative reverse transcriptase PCR, northern analysis, real-time PCR, in situ hybridization and RNase protection. dynamic range
Dynamic range as used herein is the ratio between the smallest and largest possible values of a changeable quantity. expression profile "Expression profile" as used herein may mean a genomic expression profile, e.g., an expression profile of microRNAs. Profiles may be generated by any convenient means for determining a level of a nucleic acid sequence e.g. quantitative hybridization of microRNA, labeled microRNA, amplified microRNA, cRNA, etc., quantitative PCR, ELISA for quantitation, and the like, and allow the analysis of differential gene expression between two samples. A subject or patient tumor sample, e.g., cells or collections thereof, e.g., tissues, is assayed. Samples are collected by any convenient method, as known in the art. Nucleic acid sequences of interest are nucleic acid sequences that are found to be predictive, including the nucleic acid sequences provided above, where the expression profile may include expression data for 5, 10, 20, 25, 50, 100 or more of, including all of the listed nucleic acid sequences. The term "expression profile" may also mean measuring the abundance of the nucleic acid sequences in the measured samples.
expression ratio
"Expression ratio" as used herein refers to relative expression levels of two or more nucleic acids as determined by detecting the relative expression levels of the corresponding nucleic acids in a biological sample. fragment
"Fragment" is used herein to indicate a non-full length part of a nucleic acid or polypeptide. Thus, a fragment is itself also a nucleic acid or polypeptide, respectively.
forward primer As used herein, forward primer is the sense direction primer complementary to the antisense strand of the amplicon. The forward primer may include additional nucleotides at the 5' end, that are not complement to the target sequence, thereby raising the Tm of the annealing of the primer to the target sequence. The sequence of the forward primers of the invention may comprise SEQ ED NOS: 2, 4, 6, 8, 12, 14, 16, 18, 20, 22, 40-47, 52, 55, 57, 58, 63, 64 and 69-87. gene
"Gene" as used herein may be a natural (e.g., genomic) or synthetic gene comprising transcriptional and/or translational regulatory sequences and/or a coding region and/or non- translated sequences (e.g., introns, 5'- and 3 '-untranslated sequences). The coding region of a gene may be a nucleotide sequence coding for an amino acid sequence or a functional RNA, such as tRNA, rRNA, catalytic RNA, siRNA, miRNA or antisense RNA. A gene may also be an niRNA or cDNA corresponding to the coding regions (e.g., exons and miRNA) optionally comprising 5'- or 3 '-untranslated sequences linked thereto. A gene may also be an amplified nucleic acid molecule produced in vitro comprising all or a part of the coding region and/or 5'- or 3 '-untranslated sequences linked thereto. general probe (also: universal probe, generic probe)
As used herein, a general probe binds to common sequence located in all target sequences, unspecific to the detected sequence. This is opposed to a specific probe. A general probe may be used in the detection of multiple target sequences. The sequence of the general probes of the invention may comprise SEQ. ID NOS: 48-50. groove binder/minor groove binder (MGB)
"Groove binder" and/or "minor groove binder" may be used interchangeably and refer to small molecules that fit into the minor groove of double-stranded DNA. Minor
groove binders may be long, flat molecules that can adopt a crescent-like shape and thus, fit snugly into the minor groove of a double helix, often displacing water. Minor groove binding molecules may typically comprise several aromatic rings connected by bonds with torsional freedom such as furan, benzene, or pyrrole rings. Minor groove binders may be antibiotics such as netropsin, distamycin, berenil, pentamidine and other aromatic diamidines, Hoechst 33258, SN 6999, aureolic anti-tumor drugs such as chromomycin and mithramycin, CC-1065, dihydrocyclopyrroloindole tripeptide (DPI3), l,2-dihydro-(3H)- pyrrolo[3,2-e]indole-7-carboxylate (CDPI3), and related compounds and analogues, including those described in Nucleic Acids in Chemistry and Biology, 2d ed., Blackburn and Gait, eds., Oxford University Press, 1996, and PCT Published Application No. WO 03/078450, the contents of which are incorporated herein by reference. A minor groove binder may be a component of a primer, a probe, a hybridization tag complement, or combinations thereof. Minor groove binders may increase the Tm of the primer or a probe to which they are attached, allowing such primers or probes to effectively hybridize at higher temperatures. identity
"Identical" or "identity" as used herein in the context of two or more nucleic acids or polypeptide sequences mean that the sequences have a specified percentage of residues that are the same over a specified region. The percentage may be calculated by optimally aligning the two sequences, comparing the two sequences over the specified region, determining the number of positions at which the identical residue occurs in both sequences to yield the number of matched positions, dividing the number of matched positions by the total number of positions in the specified region, and multiplying the result by 100 to yield the percentage of sequence identity. In cases where the two sequences are of different lengths or the alignment produces one or more staggered ends and the specified region of comparison includes only a single sequence, the residues of the single sequence are included in the denominator but not the numerator of the calculation. When comparing DNA and RNA, thymine (T) and uracil (U) may be considered equivalent. Identity may be performed manually or by using a computer sequence algorithm such as BLAST or BLAST 2.0. inhibit
"Inhibit" as used herein may mean prevent, suppress, repress, reduce or eliminate. isolated Nucleic Acid
The term "isolated", when used in reference to nucleic acids, is intended to mean that a nucleic acid molecule is present in a form that is substantially separated from other
naturally occurring nucleic acids that are normally associated with the molecule. Specifically, since a naturally existing chromosome (or a viral equivalent thereof) includes a long nucleic acid sequence, an "isolated nucleic acid" as used herein means a nucleic acid molecule having only a portion of the nucleic acid sequence in the chromosome but not one or more other portions present on the same chromosome. More specifically, an "isolated nucleic acid" typically includes no more than 25 kb naturally occurring nucleic acid sequences which immediately flank the nucleic acid in the naturally existing chromosome (or a viral equivalent thereof). However, it is noted that an "isolated nucleic acid" as used herein is distinct from a clone in a conventional library such as genomic DNA library and cDNA library in that the clone in a library is still in admixture with almost all the other nucleic acids of a chromosome or cell. Thus, an "isolated nucleic acid" as used herein also should be substantially separated from other naturally occurring nucleic acids that are on a different chromosome of the same organism. Specifically, an "isolated nucleic acid" means a composition in which the specified nucleic acid molecule is significantly enriched so as to constitute at least 10% of the total nucleic acids in the composition.
An "isolated nucleic acid" can be a hybrid nucleic acid having the specified nucleic acid molecule covalently linked to one or more nucleic acid molecules that are not the nucleic acids naturally flanking the specified nucleic acid. For example, an isolated nucleic acid can be in a vector. In addition, the specified nucleic acid may have a nucleotide sequence that is identical to a naturally occurring nucleic acid or a modified form or mutant thereof having one or more mutations such as nucleotide substitution, deletion/insertion, inversion, and the like. label
"Label" as used herein means a composition detectable by spectroscopic, photochemical, biochemical, immunochemical, chemical, or other physical means. For example, useful labels include 32P, fluorescent dyes, electron-dense reagents, enzymes (e.g., as commonly used in an ELISA), biotin, digoxigenin, or haptens and other entities which can be made detectable. A label may be incorporated into nucleic acids and proteins at any position. locked nucleic acid (LNA)
Locked nucleic acids as referred to herein, are modified RNA nucleotides, used to increase the sensitivity and specificity of expression in DNA molecular biology techniques based on oligonucleotides. The ribose moiety of an LNA nucleotide is modified with an
extra bridge connecting the 2' oxygen and 4' carbon. The bridge "locks" the ribose in the 3'- endo (North) conformation, which is often found in the A-form of DNA or RNA. LNA nucleotides can be mixed with DNA or KNA bases in the oligonucleotide when desired. The locked ribose conformation enhances base stacking and backbone pre-organization, thereby increasing the thermal stability (melting temperature) of oligonucleotides. mismatch
"Mismatch" means a nucleobase of a first nucleic acid that is not capable of pairing with a nucleobase at a corresponding position of a second nucleic acid. modulation "Modulation" as used herein means a perturbation of function or activity, hi certain embodiments, modulation means an increase in gene expression, m certain embodiments, modulation means a decrease in gene expression. nucleic acid "Nucleic acid" or "oligonucleotide" or "polynucleotide" as used herein mean at least two nucleotides covalently linked together. The depiction of a single strand also defines the sequence of the complementary strand. Thus, a nucleic acid also encompasses the complementary strand of a depicted single strand. Many variants of a nucleic acid may be used for the same purpose as a given nucleic acid. Thus, a nucleic acid also encompasses substantially identical nucleic acids and complements thereof. A single strand provides a probe that may hybridize to a target sequence under stringent hybridization conditions. Thus, a nucleic acid also encompasses a probe that hybridizes under stringent hybridization conditions.
Nucleic acids may be single stranded or double stranded, or may contain portions of both double stranded and single stranded sequence. The nucleic acid may be DNA, both genomic and cDNA, RNA, or a hybrid, where the nucleic acid may contain combinations of deoxyribo- and ribo-nucleotides, and combinations of bases including uracil, adenine, thymine, cytosine, guanine, inosine, xanthine hypoxanthine, isocytosine and isoguanine.
Nucleic acids may be obtained by chemical synthesis methods or by recombinant methods.
A nucleic acid will generally contain phosphodiester bonds, although nucleic acid analogs may be included that may have at least one different linkage, e.g., phosphoramidate, phosphorothioate, phosphorodithioate, or O-methylphosphoroamidite linkages and peptide nucleic acid backbones and linkages. Other analog nucleic acids include those with positive backbones; non-ionic backbones, and non-ribose backbones, including those described in
U.S. Pat. Nos. 5,235,033 and 5,034,506, which are incorporated by reference. Nucleic acids containing one or more non-naturally occurring or modified nucleotides are also included within one definition of nucleic acids. The modified nucleotide analog may be located for example at the 5'-end and/or the 3'-end of the nucleic acid molecule. Representative examples of nucleotide analogs may be selected from sugar- or backbone-modified ribonucleotides. It should be noted, however, that also nucleobase-modified ribonucleotides, i.e. ribonucleotides, containing a non-naturally occurring nucleobase instead of a naturally occurring nucleobase such as uridines or cytidines modified at the 5-position, e.g. 5-(2- amino) propyl uridine, 5-bromo uridine; adenosines and guanosines modified at the 8- position, e.g. 8-bromo guanosine; deaza nucleotides, e.g. 7-deaza-adenosine; O- and N- alkylated nucleotides, e.g. N6-methyl adenosine are suitable. The 2'-OH-group may be replaced by a group selected from H5 OR, R, halo, SH, SR, NH2, NHR, NR2 or CN, wherein R is C1-C6 alkyl, alkenyl or alkynyl and halo is F5 Cl, Br or I. Modified nucleotides also include nucleotides conjugated with cholesterol through, e.g., a hydroxyprolinol linkage as described in Krutzfeldt et al, Nature 438:685-689 (2005) and Soutschek et al., Nature 432:173-178 (2004), which are incorporated herein by reference. Modifications of the ribose-phosphate backbone may be done for a variety of reasons, e.g., to increase the stability and half-life of such molecules in physiological environments, to enhance diffusion across cell membranes, or as probes on a biochip. The backbone modification may also enhance resistance to degradation, such as in the harsh endocytic environment of cells. The backbone modification may also reduce nucleic acid clearance by hepatocytes, such as in the liver. Mixtures of naturally occurring nucleic acids and analogs may be made; alternatively, mixtures of different nucleic acid analogs, and mixtures of naturally occurring nucleic acids and analogs may be made. partially identical
" partially identical" as used herein means that a first sequence is at least 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 97%, 98% or 99% identical to a second sequence over a region of 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70, 75, 80, 85, 90, 95, 100 or more nucleotides, or that the two sequences hybridize under stringent hybridization conditions. polyadenylation
As used herein "Polyadenylation" is the addition of a poly(A) tail to an RNA molecule. The poly(A) tail is a stretch of RNA consisting of multiple adenosine monophosphates.
primer
As used herein, a primer is a strand of nucleic acid that serves as a starting point for DNA replication. They are required because DNA polymerases can only add new nucleotides to an existing strand of DNA. The polymerase starts replication at the 3'-end of the primer, and copies the opposite strand.
Laboratory techniques that involve DNA polymerase, such as polymerase chain reaction (PCR), require DNA primers. These primers are usually short, chemically synthesized oligonucleotides. They are hybridized to a target DNA, which is then copied by the polymerase. primer-dimer
A primer-dimer as used herein means a potential by-product in PCR. Primer-dimers consist of primer molecules that have hybridized to each other due to complementary bases in the primers. As a result, the DNA polymerase amplifies the primer-dimer, leading to competition for PCR reagents, thus potentially inhibiting amplification of the DNA sequence targeted for PCR amplification. In real-time PCR, primer-dimer s may interfere with accurate quantification. probe
"Probe" as used herein means an oligonucleotide capable of binding to a target nucleic acid of complementary sequence through one or more types of chemical bonds, usually through complementary base pairing, usually through hydrogen bond formation.
Probes may bind target sequences lacking complete complementarity with the probe sequence depending upon the stringency of the hybridization conditions. There may be any number of base pair mismatches which will interfere with hybridization between the target sequence and the single stranded nucleic acids described herein. However, if the number of mismatches is so great that no hybridization can occur under even the least stringent of hybridization conditions, the sequence is not a complementary target sequence. A probe may be single stranded or partially single and partially double stranded. The strandedness of the probe is dictated by the structure, composition, and properties of the target sequence.
Probes may be directly labeled or indirectly labeled. An exemplary probe is TaqMan MGB probe that contains 5' fluorescence, a 3' quencher arid MGB. promoter
"Promoter" as used herein means a synthetic or naturally-derived molecule which is capable of conferring, activating or enhancing expression of a nucleic acid in a cell. A
promoter may comprise one or more specific transcriptional regulatory sequences to further enhance expression and/or to alter the spatial expression and/or temporal expression of same. A promoter may also comprise distal enhancer or repressor elements, which can be located as much as several thousand base pairs from the start site of transcription. A promoter may be derived from sources including viral, bacterial, fungal, plants, insects, and animals. A promoter may regulate the expression of a gene component constitutively, or differentially with respect to cell, the tissue or organ in which expression occurs or, with respect to the developmental stage at which expression occurs, or in response to external stimuli such as physiological stresses, pathogens, metal ions, or inducing agents. Representative examples of promoters include the bacteriophage T7 promoter, bacteriophage T3 promoter, SP6 promoter, lac operator-promoter, tac promoter, SV40 late promoter, SV40 early promoter, RSV-LTR promoter, CMV IE promoter, SV40 early promoter or S V40 late promoter and the CMV IE promoter. quencher A label may also be a quencher molecule, which when in proximity to another label, may decrease the amount of detectable signal of the other label, such as described in U.S. Patent No. 6,541,618, the contents of which are incorporated herein by reference. A label may be incorporated into nucleic acids and proteins at any position. reverse primer As used herein, the primer complementary to a sense polyadenylated strand is designated the reverse primer. The sequence of the reverse primer of the invention may comprise SEQ. ID NO: 28. selectable marker
"Selectable marker" as used herein means any gene which confers a phenotype on a host cell in which it is expressed to facilitate the identification and/or selection of cells which are transfected or transformed with a genetic construct. Representative examples of selectable markers include the ampicillin-resistance gene (Amp1), tetracycline-resistance gene (Tc1), bacterial kanamycin-resistance gene (Kan1), zeocin resistance gene, the AURI-C gene which confers resistance to the antibiotic aureobasidin A, phosphinothricin-resistance gene, neomycin phosphotransferase gene (nptU), hygromycin-resistance gene, beta- glucuronidase (GUS) gene, chloramphenicol acetyltransferase (CAT) gene, green fluorescent protein (GFP)-encoding gene and luciferase gene.
sense probe
The term "sense probe" as used herein, refers to a probe which is complementary and hybridizes to the universal region on the antisense strand (- strand) of the amplicon generated by RT-PCR. The sequence of the sense probe of the invention may comprise the sequence of SEQ. ID NO: 49. stringent hybridization conditions
"Stringent hybridization conditions" as used herein mean conditions under which a first nucleic acid sequence (e.g., probe) will hybridize to a second nucleic acid sequence
(e.g., target), such as in a complex mixture of nucleic acids. Stringent conditions are sequence-dependent and will be different in different circumstances. Stringent conditions may be selected to be about 5-1O0C lower than the thermal melting point (Tm) for the specific sequence at a defined ionic strength pH. The Tm may be the temperature (under defined ionic strength, pH, and nucleic acid concentration) at which 50% of the probes complementary to the target hybridize to the target sequence at equilibrium (as the target sequences are present in excess, at Tm, 50% of the probes are occupied at equilibrium).
Stringent conditions may be those in which the salt concentration is less than about
1.0 M sodium ion, such as about 0.01-1.0 M sodium ion concentration (or other salts) at pH
7.0 to 8.3 and the temperature is at least about 3O0C for short probes (e.g., about 10-50 nucleotides) and at least about 6O0C for long probes (e.g., greater than about 50 nucleotides). Stringent conditions may also be achieved with the addition of destabilizing agents such as formamide. For selective or specific hybridization, a positive signal may be at least 2 to 10 times background hybridization. Exemplary stringent hybridization conditions include the following: 50% formamide, 5x SSC, and 1% SDS, incubating at
42°C, or, 5x SSC, 1% SDS, incubating at 65°C, with wash in 0.2x SSC, and 0.1% SDS at 650C. substantially complementary
"Substantially complementary" as used herein means that a first sequence is at least 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 97%, 98% or 99% identical to the complement of a second sequence over a region of 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70, 75, 80, 85, 90, 95, 100 or more nucleotides, or that the two sequences hybridize under stringent hybridization conditions. substantially identical "Substantially identical" as used herein means that a first and a second sequence are at least 60%, 65%, 70%, 75%, 80%, 85%, 90%, 95%, 97%, 98% or 99% identical over a
region of 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 30, 35, 40, 45, 50, 55, 60, 65, 70, 75, 80, 85, 90, 95, 100 or more nucleotides or amino acids, or with respect to nucleic acids, if the first sequence is substantially complementary to the complement of the second sequence. target nucleic acid
"Target nucleic acid" as used herein means a nucleic acid or variant thereof that may be bound by another nucleic acid. A target nucleic acid may be a DNA sequence. The target nucleic acid may be RNA. The target nucleic acid may comprise an rnRNA, tRNA, shRNA, siRNA or Piwi-interacting RNA, or a pri-miRNA, pre-miRNA, miRNA, or anti-miRNA. One or more probes may bind the target nucleic acid. The target binding site may comprise 5-100 or 10-60 nucleotides. The target binding site may comprise a total of 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15,16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30-40, 40- 50, 50-60, 61, 62 or 63 nucleotides. tissue sample As used herein, a tissue sample is tissue obtained from a tissue biopsy using methods well known to those of ordinary skill in the related medical arts. The phrase "suspected of being cancerous" as used herein means a cancer tissue sample believed by one of ordinary skill in the medical arts to contain cancerous cells. Methods for obtaining the sample from the biopsy include gross apportioning of a mass, microdissection, laser-based microdissection, or other art-known cell-separation methods. universal adaptor
As used herein, a universal adaptor is not specific to the amplified target. universal primer
As used herein, a universal primer is not unique to a specific sequence targeted for amplification, and may hybridize to all sequences comprising a common region complementary to the primer. This is opposed to a specific primer which is specific to the amplified sequence. A universal primer may be used in the amplification of multiple target sequences. variant "Variant" as used herein referring to a nucleic acid means (i) a portion of a referenced nucleotide sequence; (ii) the complement of a referenced nucleotide sequence or portion thereof; (iii) a nucleic acid that is substantially identical to a referenced nucleic acid or the
complement thereof; or (iv) a nucleic acid that hybridizes under stringent conditions to the referenced nucleic acid, complement thereof, or a sequence substantially identical thereto. vector
"Vector" as used herein means a nucleic acid sequence containing an origin of replication. A vector may be a plasmid, bacteriophage, bacterial artificial chromosome or yeast artificial chromosome. A vector may be a DNA or RNA vector. A vector may be either a self-replicating extrachromosomal vector or a vector which integrates into a host genome. wild type As used herein, the term "wild type" sequence refers to a coding, a non-coding or an interface sequence which is an allelic form of sequence that performs the natural or normal function for that sequence. Wild type sequences include multiple allelic forms of a cognate sequence, for example, multiple alleles of a wild type sequence may encode silent or conservative changes to the protein sequence that a coding sequence encodes.
2. MicroRNAs and their processing
A gene coding for a microRNA (miRNA) may be transcribed leading to production of an miRNA precursor known as the pri-miRNA. The pri-miRNA may be part of a polycistronic RNA comprising multiple pri-miRNAs. The pri-miRNA may form a hairpin structure with a stem and loop. The stem may comprise mismatched bases.
The hairpin structure of the pri-miRNA may be recognized by Drosha, which is an RNase III endonuclease. Drosha may recognize terminal loops in the pri-miRNA and cleave approximately two helical turns into the stem to produce a 60-70 nucleotide precursor known as the pre-miRNA. Drosha may cleave the pri-miRNA with a staggered cut typical of RNase III endonucleases yielding a pre-miRNA stem loop with a 5' phosphate and ~2 nucleotide 3' overhang. Approximately one helical turn of the stem (~10 nucleotides) extending beyond the Drosha cleavage site may be essential for efficient processing. The pre-miRNA may then be actively transported from the nucleus to the cytoplasm by Ran-GTP and the export receptor Ex-portin-5. The pre-miRNA may be recognized by Dicer, which is also an RNase III endonuclease. Dicer may recognize the double-stranded stem of the pre-miRNA. Dicer may also recognize the 5' phosphate and 3' overhang at the base of the stem loop. Dicer may cleave off the terminal loop two helical turns away from the base of the stem loop
leaving an additional 5' phosphate and ~2 nucleotide 3' overhang. The resulting siRNA-like duplex, which may comprise mismatches, comprises the mature miRNA and a similar-sized fragment known as the miRNA*. The miRNA and miRNA* may be derived from opposing arms of the pri-miRNA and pre-miRNA. MiRNA* sequences may be found in libraries of cloned miRNAs but typically at lower frequency than the miRNAs.
Although initially present as. a double-stranded species with miRNA*, the miRNA may eventually become incorporated as a single-stranded RNA into a ribonucleoprotein complex known as the RNA-induced silencing complex (RISC). Various proteins can form the RISC, which can lead to variability in specificity for miRNA/miRNA* duplexes, binding site of the target gene, activity of miRNA (repression or activation), and which strand of the miRNA/miRNA* duplex is loaded in to the RISC.
When the miRNA strand of the miRNA:miRNA* duplex is loaded into the RISC, the miRNA* may be removed and degraded. The strand of the miRNA:miRNA* duplex that is loaded into the RISC may be the strand whose 5' end is less tightly paired, hi cases where both ends of the miRNA:miRNA* have roughly equivalent 5' pairing, both miRNA and miRNA* may have gene silencing activity.
The RISC may identify target nucleic acids based on high levels of complementarity between the miRNA and the mRNA, especially by nucleotides 2-7 of the miRNA. Only one case has been reported in animals where the interaction between the miRNA and its target was along the entire length of the miRNA. This was shown for mir-196 and Hox B 8 and it was further shown that mir-196 mediates the cleavage of the Hox B8 mRNA (Yekta et al 2004, Science 304-594). Otherwise, such interactions are known only in plants (Bartel & Bartel 2003, Plant Physiol 132-709).
A number of studies have studied the base-pairing requirement between miRNA and its mRNA target for achieving efficient inhibition of translation (reviewed by Bartel 2004, Cell 116-281). In mammalian cells, the first 8 nucleotides of the miRNA may be important (Doench & Sharp 2004 GenesDev 2004-504). However, other parts of the microRNA may also participate in mRNA binding. Moreover, sufficient base pairing at the 3' can compensate for insufficient pairing at the 5' (Brennecke et al, 2005 PLoS 3-e85). Computation studies, analyzing miRNA binding on whole genomes have suggested a specific role for bases 2-7 at the 5' of the miRNA in target binding but the role of the first nucleotide, found usually to be "A" was also recognized (Lewis et at 2005 Cell 120-15). Similarly, nucleotides 1-7 or 2-8 were used to identify and validate targets by Krek et al (2005, Nat Genet 37-495).
The target sites in the niRNA may be in the 5' UTR, the 3' UTR or in the coding region. Interestingly, multiple miRNAs may regulate the same mRNA target by recognizing the same or multiple sites. The presence of multiple miRNA binding sites in most genetically identified targets may indicate that the cooperative action of multiple RISCs provides the most efficient translational inhibition. miRNAs may direct the RISC to downregulate gene expression by either of two mechanisms: mRNA cleavage or translational repression. The miRNA may specify cleavage of the mRNA if the mRNA has a certain degree of complementarity to the miRNA. When a miRNA guides cleavage, the cut may be between the nucleotides pairing to residues 10 and 11 of the miRNA. Alternatively, the miRNA may repress translation if the miRNA does not have the requisite degree of complementarity to the miRNA. Translational repression may be more prevalent in animals since animals may have a lower degree of complementarity between the miRNA and the binding site.
It should be noted that there may be variability in the 5' and 3' ends of any pair of miRNA and miRNA* . This variability may be due to variability in the enzymatic processing of Drosha and Dicer with respect to the site of cleavage. Variability at the 5' and 3' ends of miRNA and miRNA* may also be due to mismatches in the stem structures of the pri- miRNA and pre-miRNA. The mismatches of the stem strands may lead to a population of different hairpin structures. Variability in the stem structures may also lead to variability in the products of cleavage by Drosha and Dicer.
3. Nucleic Acids
Nucleic acids are provided herein. The nucleic acids of the invention comprise the sequence of the provided nucleic acids or variants thereof. The variant may be a complement of the referenced nucleotide sequence. The variant may also be a nucleotide sequence that is substantially identical to the referenced nucleotide sequence or the complement thereof. The variant may also be a nucleotide sequence which hybridizes under stringent conditions to the referenced nucleotide sequence, complements thereof, or nucleotide sequences substantially identical thereto.
The nucleic acid may have a length of from 10 to 250 nucleotides. The nucleic acid may have a length of at least 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 35, 40, 45, 50, 60, 70, 80, 90, 100, 125, 150, 175, 200 or 250 nucleotides. The nucleic acid may be synthesized or expressed in a cell (in vitro or in vivo) using a synthetic gene described herein. The nucleic acid may be synthesized as a single strand molecule and hybridized to a substantially complementary nucleic acid to form a duplex.
The nucleic acid may be introduced to a cell, tissue or organ in a single- or double-stranded form or capable of being expressed by a synthetic gene using methods well known to those skilled in the art, including as described in U.S. Patent No. 6,506,559 which is incorporated by reference. 3a. Nucleic acid complexes
The nucleic acid may further comprise one or more of the following: a peptide, a protein, a RNA-DNA hybrid, an antibody, an antibody fragment, a Fab fragment, and an aptamer.
3b. Pri-miRNA The nucleic acid may comprise a sequence of a pri-miRNA or a variant thereof. The pri-miRNA sequence may comprise from 45-30,000, 50-25,000, 100-20,000, 1,000-1,500 or 80-100 nucleotides. The sequence of the pri-miRNA may comprise a pre-miRNA, miRNA and miRNA*, as set forth herein, and variants thereof.
The pri-miRNA may form a hairpin structure. The hairpin may comprise a first and a second nucleic acid sequence that are substantially complimentary. The first and second nucleic acid sequence may be from 37-50 nucleotides. The first and second nucleic acid sequence may be separated by a third sequence of from 8-12 nucleotides. The hairpin structure may have a free energy of less than -25 Kcal/mole, as calculated by the Vienna algorithm, with default parameters as described in Hofacker et al., Monatshefte f. Chemie 125: 167-188 (1994), the contents of which are incorporated herein. The hairpin may comprise a terminal loop of 4-20, 8-12 or 10 nucleotides. The pri-miRNA may comprise at least 19% adenosine nucleotides, at least 16% cytosine nucleotides, at least 23% thymine nucleotides and at least 19% guanine nucleotides. 3c. Pre-miRNA The nucleic acid may also comprise a sequence of a pre-miRNA or a variant thereof.
The pre-miRNA sequence may comprise from 45-90, 60-80 or 60-70 nucleotides. The sequence of the pre-miRNA may comprise a miRNA and a miRNA* as set forth herein. The sequence of the pre-miRNA may also be that of a pri-miRNA excluding from 0-160 nucleotides from the 5' and 3' ends of the pri-miRNA. 3d. miRNA
The nucleic acid may also comprise a sequence of a miRNA (including miRNA*) or a variant thereof. The miRNA sequence may comprise from 13-33, 18-24 or 21-23 nucleotides. The miRNA may also comprise a total of at least 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37,
38, 39 or 40 nucleotides. The sequence of the miRNA may be the first 13-33 nucleotides of the pre-miRNA. The sequence of the miRNA may also be the last 13-33 nucleotides of the pre-miRNA.
3e. Anti-miRNA The nucleic acid may also comprise a sequence of an anti-miRNA capable of blocking the activity of a miRNA or miRNA*, such as by binding to the pri-miRNA, pre- miRNA, miRNA or miRNA* (e.g. antisense or RNA silencing), or by binding to the target binding site. The anti-miRNA may comprise a total of 5-100 or 10-60 nucleotides. The anti-miRNA may also comprise a total of at least 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38, 39 or 40 nucleotides. The sequence of the anti-miRNA may comprise (a) at least 5 nucleotides that are substantially identical or complimentary to the 5' of a miRNA and at least 5-12 nucleotides that are substantially complimentary to the flanking regions of the target site from the 5' end of the miRNA, or (b) at least 5-12 nucleotides that are substantially identical or complimentary to the 3' of a miRNA and at least 5 nucleotide that are substantially complimentary to the flanking region of the target site from the 3' end of the miRNA.
3f. Binding Site of Target
The nucleic acid may also comprise a sequence of a target microRNA binding site or a variant thereof. The target site sequence may comprise a total of 5-100 or 10-60 nucleotides.
The target site sequence may also comprise a total of at least 5, 6, 7, 8, 9, 10, 11, 12, 13, 14,
15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34, 35, 36, 37, 38,
39, 40, 41, 42, 43, 44, 45, 46, 47, 48, 49, 50, 51, 52, 53, 54, 55, 56, 57, 58, 59, 60, 61, 62 or 63 nucleotides. 4. Probes
A probe is provided herein. A probe may comprise a nucleic acid. The probe may have a length of from 8 to 500, 10 to 100 or 20 to 60 nucleotides. The probe may also have a length of at least 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 35, 40, 45, 50, 60, 70, 80, 90, 100, 120, 140, 160, 180, 200, 220, 240, 260, 280 or 300 nucleotides. The probe may comprise a nucleic acid of 18-25 nucleotides. The sequence of the probes of the invention may comprise the sequence of SEQ ID NOS: 48-50 or variants thereof.
A probe may be capable of binding to a target nucleic acid of complementary sequence through one or more types of chemical bonds, usually through complementary
base pairing, usually through hydrogen bond formation. Probes may bind target sequences lacking complete complementarity with the probe sequence depending upon the stringency of the hybridization conditions. A probe may be single stranded or partially single and partially double stranded. The strandedness of the probe is dictated by the structure, composition, and properties of the target sequence. Probes may be directly labeled or indirectly labeled.
The probe may further comprise a linker. The linker may be 10-60 nucleotides in length. The linker may be 20-27 nucleotides in length. The linker may be of sufficient length to allow the probe to be a total length of 45-60 nucleotides. The linker may not be capable of forming a stable secondary structure, or may not be capable of folding on itself, or may not be capable of folding on a non-linker portion of a nucleic acid contained in the probe.
5. Reverse Transcription
Target sequences of a cDNA may be generated by reverse transcription of the target KNA. Methods for generating cDNA may be reverse transcribing polyadenylated RNA or alternatively, RNA with a ligated adaptor sequence.
The RNA may be ligated to an adaptor sequence prior to reverse transcription. A ligation reaction may be performed by T4 RNA ligase to ligate an adaptor sequence at the 3' end of the RNA. Reverse transcription (RT) reaction may then be performed using a primer comprising a sequence that is reverse complementary to the 3' end of the adaptor sequence.
Polyadenylated RNA may be used in a reverse transcription (RT) reaction using a poly(T) primer (also referred to as oligo dT) comprising a 5' adaptor sequence. The poly(T) sequence may comprise 8, 9, 10, 11, 12, 13, or 14 consecutive thymines.
The reverse transcript of the RNA may be amplified by real time PCR, using a specific forward primer comprising at least 15 nucleic acids complementary to the target nucleic acid and a 5' tail sequence; a reverse primer that is complementary to the 3' end of the adaptor sequence; and a probe that may be specifically complementary to the target nucleic acid. The probe may be partially complementary to the 5' end of the adaptor sequence. Methods of amplifying target nucleic acids are described herein. The amplification may be by a method comprising PCR. The first cycles of the PCR reaction may have an annealing temp of 56°C, 57°C, 58°C, 59°C, or 60°C. The first cycles may comprise 1-10 cycles. The remaining cycles of the PCR reaction may be 6O0C. The remaining cycles may
comprise 2-40 cycles. The annealing temperature may cause the PCR to be more sensitive. The PCR may generate longer products that can serve as higher stringency PCR templates.
The PCR reaction may comprise a forward primer. The forward primer may comprise 15, 16, 17, 18, 19, 20, or 21 nucleotides identical to the target nucleic acid. The 3' end of the forward primer may be sensitive to differences in sequence between a target nucleic acid and a sibling nucleic acid.
The forward primer may also comprise a 5' overhanging tail. The 5' tail may increase the melting temperature of the forward primer. The sequence of the 5' tail may comprise a sequence that is non-identical to the genome of the animal from which the target nucleic acid is isolated. The sequence of the 5' tail may also be synthetic. The 5' tail may comprise 8, 9, 10, 11, 12, 13, 14, 15, or 16 nucleotides.
The PCR reaction may comprise a reverse primer. The reverse primer may be complementary to a target nucleic acid. The reverse primer may also comprise a sequence complementary to an adaptor sequence. The sequence complementary to an adaptor sequence may comprise 12-24 nucleotides.
6. Kits
A kit is also provided and may comprise a nucleic acid described herein together with any or all of the following: assay reagents, buffers, probes and/or primers, and sterile saline or another pharmaceutically acceptable emulsion and suspension base. In addition, the kits may include instructional materials containing directions (e.g., protocols) for the practice of the methods described herein.
For example, the kit may be used for the amplification, detection, identification or quantification of a target nucleic acid sequence. The kit may comprise a poly(T) primer, a forward primer, a reverse primer, and a probe. Any of the compositions described herein may be comprised in a kit. In a non- limiting example, reagents for isolating miRNA, labeling miRNA, and/or evaluating a miRNA population using an array are included in a kit. The kit may further include reagents for creating or synthesizing miRNA probes. The kits will thus comprise, in suitable container means, an enzyme for labeling the miRNA by incorporating labeled nucleotide or unlabeled nucleotides that are subsequently labeled. It may also include one or more buffers, such as reaction buffer, labeling buffer, washing buffer, or a hybridization buffer, compounds for preparing the miRNA probes, components for in situ hybridization and components for isolating miRNA. Other kits of the invention may include components for
making a nucleic acid array comprising miRNA, and thus, may include, for example, a solid support.
The following examples are presented in order to more fully illustrate some embodiments of the invention. They should, in no way be construed, however, as limiting the broad scope of the invention.
EXAMPLES
Example 1: miRNA detection by the General Probe Detection assay, SYBRGreen, and Varkonyi-Gasic (stem-loop) techniques hsa-miR-296-3ρ (SEQ. ID NO: 11) was detected by the General (antisense) Probe
Detection Assay of the invention, and by the SYBR Green method; hsa-miR-181a (SEQ. ID
NO: 7) was detected by the General (antisense) Probe Detection Assay of the invention, and by the Varkonyi-Gasic (stem-loop) method. The results of each pair of detection methods were compared.
Example 1.1 Materials and methods 1.1.1 Cells and viruses :
Human cell lines UKF-NB4 (neuroblastoma) and B95-8, that harbors Epstein-Barr Virus (EBV), were cultured in RPMI- 1640 Medium supplemented with 10% heat inactivated fetal calf serum (FCS), 1% penicillin-streptomycin and 1% L-glutamine (Biological Industries, Beit Haemek, IS), at 37°C in a 5% CO2 atmosphere.
1.1.2. RNA isolation and cDNA preparation, and Real-time PCR ("RTPCR) reactions
Total RNA was isolated using EZ-RNA isolation kit (Biological Industries, Beit Haemek, Israel), according to the manufacturer's protocol. All real-time PCR reactions were conducted on light cycler 480® (Roche
Diagnostics, Manheim, Germany), in 96-well plates. All primers and synthetic miRNA oligonucleotides were purchased from Integrated DNA Technologies (IDT, Coralville, IA), except for the UPL#61 probe (SEQ. ID NO: 50) (Roche diagnostics), a dual labeled 8 nucleotide hydrolysis probe, one of which is a LNA (locked nucleic acid). The sequences are detailed in table 1 below. Samples were run in duplicate, and each experiment was repeated three times.
1.1.3 General Probe Detection Assay analysis of miRNAs
For the cDNA preparation total RNA (1 μg) was polyadenylated in a 10 μl reaction containing 2.5 niM MnCl2, PNK buffer (New England Biolabs, Ipswich, MA), 4mM ATP (Amersham Pharmacia, Uppsala, Sweden) and 1.5 units of poly A polymerase (Takara,
Shiga, Japan), as described by Shi and Chiang, with a different RT primer (RTl) and without the precipitation stage. After one hour of incubation at 37°c, 5μl were mixed with
0.5μg RTl primer (SEQ. ID NO: 26) (the sequence of this RTl primer is composed of a 3' rapid amplification of complementary DNA ends tail, and the UPL#61 sequence). The reaction was heated to 85°c for 2 min in order to disrupt any existing RNA secondary structures, followed by temperature gradient in which the temperature decreased gradually from 75°c to 25°c. Each sample was then combined with 12μl of RT mix, to a final volume of 20μl, containing 0.125mM of each dNTP (Amersham, Uppsala, Sweden), 0.255M trealose, 5x RT buffer, 1OmM DTT and 200 units of Superscript II reverse transcriptase (Invitrogen, Carlsbad, CA). For each sample a control which did not contain reverse transcriptase was prepared (further referred to as minus-RT control). The reaction was heated for 5 min at 37°c and 5 min at 45°c for 5 cycles.
The real-time PCR reactions of the General Probe Detection Assay were performed in lOμl volume reaction which included 5μl ABsolute™ qPCR mix (ABgene, IL, USA), 0.5μM miR-specific forward primer (SEQ. ID NO: 12, for hsa-miR-296-3ρ, and SEQ. ID NO: 8, for hsa-miR-181a), 0.5μM reverse primer (SEQ. ID NO: 28), and 0.05μM UPL#61 (SEQ. ID NO: 50). The reactions were incubated at 95°c for 15 min followed by 45 cycles of 95°c for 15s, 60°c for 20s and 60°c for 20s, and then cooled to 40°c.
A schematic description of the General (antisense) Probe Detection assay is shown in Figure IA.
1.1.4 SYBR Green analysis of miRNAs
PoIyA based analysis of miRNAs was done using SYBR Green as described above except for the use of a different RT primer, RT2 (SEQ. ID NO: 27), which lacks the UPL#61 binding sequence. The real-time PCR reactions of the SYBR Green analysis was performed in a 10 μl volume reaction which included 2X Power SYBR® Green PCR master mix (Applied Biosystems, Foster City, CA), 0.5μM forward hsa-miR-296-3p specific primer (SEQ. ID NO: 12) and 0.5 μM reverse primer (SEQ. ID NO: 28). The reactions were incubated at
95°c for 10 min followed by 45 cycles of 95°c for 10s and 60°c for 1 min. For melting curve analysis the reactions were heated to 95°c for 5s and 60°c for 15s. The reactions were then cooled to 400C.
1.1.5 The Varkonyi-Gasic (Stem-loop) based analysis of miRNAs
The Varkonyi-Gasic method for miRNA detection is done in one tube and does not involve cDNA preparation.
The real-time PCR reactions were conducted as follows: Stem-loop qPCR amplification of miR-181a (SEQ. TD NO: 7) was performed using RNA Ultrasense™ One- Step RT-PCR kit (hwitrogen, Carlsbad, CA) as described (Varkonyi-Gasic et al, 2007). The RT, the forward, and the reverse primers sequences are provided in table 1. UPL#61 (SEQ. ID NO: 50) was added at a final concentration of 0.05 μM. Reaction was carried out as follows: 50°c for 15 min, 95°c for 2 min, followed by 45 cycles of 95°c for 15s and 60°c for 30s.
A schematic description of the miRNA stem-loop-qRT-PCR assay is shown in Figure IB.
Table 1: Primers used in reverse transcription and qRT-PCR reactions for miRNA detection by the General Probe Detection assay, SYBR Green, and Varkonyi-Gasic (stem-loo techni ues
*V=A, G, C; N=A, T, G, C.
Example 1.2 Comparison of the detection hsa-miR-296-3p (SEQ. ID NO: 11) by the General Probe Detection assay with its detection by SYBR Green hsa-miR-296-3p (SEQ. ID NO: 11) was amplified using both of the General Probe
Detection assay and by SYBR Green.
With amplification by the SYBR-Green-polyA-based-technique, the non-template negative control gave a very clear product in the RT-PCR (Figure 2A), probably originating from SYBR Green that detected fluorescence from primer-dimers rather than a specific product. As indicated in Figure 3 A, analysis of SYBR Green amplification of hsa-miR-296-
3p (SEQ. ID NO: 11) on agarose gel could not demonstrate clear expression of this miR.
When qPCR was performed with the General (antisense) Probe Assay, it was possible to detect hsa-naiR-296-3p (SEQ. ID NO: 11) in 25ρg of cellular RNA at a crossing point (Cp) value of 34, and no signals were detected in the negative controls (no-template- control, and no-RT reactions) (Figure 2B). When the qPCR products of the General Probe Detection assay were run on 2.5% gel no pre-microRNAs were observed, thereby indicating that the detection was specific to mature miRNA (Figure 3C). The size of the mature hsa- miR-296-3ρ (SEQ. ID NO: 11) is -70 nucleotides.
Example 1.3 Comparison of the detection of hsa-miR-181a (SEQ. ID NO: 7) by the General Probe Detection assay with its detection by the Varkonyi-Gasic (stem-loop) method
As indicated in Figure 2D, in the Varkonyi-Gasic stem-loop qRT-PCR amplification of hsa-miR-181a (SEQ. ID NO: T) the no template-control gave rise to a Cp value of 35.1 whereas 25, 2.5 and 0.25 pg of cDNA gave rise to Cp values of 35.5, 37.2 and 37.5, respectively. This is also apparent from the analysis on 2.5% agarose gel in Figure 3B.
In contrast, the General (antisense) Probe miR assay detected hsa-miR-181a (SEQ. ID NO: 7) in 0.25 pg of cDNA, and no fluorescence was detected in the no-template negative control (Figure 2C). When the qPCR products of the General Probe Detection assay were run on 2.5% gel no pre-microRNAs were observed, thereby indicating that the detection was specific to mature miRNA (Figure 3C). The size of this mature miR hsa-miR- 181a (SEQ. ID NO: 7) is also -70 nucleotides, as expected.
Example 2: Specificity of the General (antisense) Probe Detection assay
Example 2.1 Cross reactivity between miRNAs within the let-7 family
A miRNA family is typically characterized by single to a few nucleotides differences within the 3'-end of its miRNAs members. The ability of the General (antisense) Probe Detection assay to discriminate miRNAs within a family was tested on the let-7 family. Two cDNA libraries were prepared from synthetic miRNA sequences of let-7c and let-7 d, and were then amplified using the sequences of hsa-let-7a to -7e as forward primers. Relative detection specificity the General (antisense) Probe Detection assay for each forward primer was calculated as a percent of the perfect match of the Cp values of target and off-target assays (100x2"ΛCp) when using 250pg of cDNA prepared from synthetic RNA.
Table 2: Primer sequences and calculated relative detection
As depicted in table 2, very low signals of non-specific amplification were observed for let-7c cDNA when it was tested with the forward primers let-7a (SEQ. ID NO: 14), let- 7b (SEQ. ID NO: 16), let-7d (SEQ. ID NO: 20), and let-7e (SEQ. ID NO: 22) (0.3%, 0.8%, 0%, and 0%, respectively). The let-7d cDNA gave similar results except for let-7a (SEQ. ID NO: 14), where the assay could detect 32 % of the target sequence, probably as a result of the wobble at the 5'end of the RT primer.
Example 2.2 Profiling expression of EBV-miRNAs
The sensitivity of the General Probe Detection assay and its ability to detect specific viral miRNAs in the presence of other host miRNAs were determined in a competition assay. One μg of total RNA isolated from B95-8 cells (a B cell line infected by EBV) and 1
μg of total RNA isolated from UKF-NB4 cells were reverse transcribed separately. The cDNA library of the B95-8 cells was serially diluted by two folds, from 250 to 0.25 pg, with the cDNA library of the UKF-NB4 cells. The diluted B-95 library was then used in the General (antisense) Probe Detection assay to detect EBV-miR-BARTl (SEQ. ID NO: 23), with the specific EBV-miR-BARTl primer (SEQ. ID NO: 24). EBV-BARTl (SEQ. ID NO: 23) could be detected in as little as 0.25 pg of cDNA (Fig. 4A) originated from B-95-8 total RNA (in total of 0.25 ng cDNA). As in the case of host miRNAs, no fluorescence was detected in the -RT control or in the no-template negative controls (Fig. 4B).
Example 2.3 Validation of miRNAs
Additional viral and human miRNAs were validated using total RNA from UKF- NB4 and EBV infected cells (B95-8) by the General (antisense) Probe Detection Assay. Details of the sequences are detailed in table 3 below.
Table 3: miRNA sequences and primers used in validation of miRNAs
The General (antisense) Probe Detection assay showed excellent linearity over five orders of magnitude ranging from 0.25 to 2500 pg. As indicated in Figure 5A, it was possible to detect hsa-miR-30a (SEQ. ID NO: 1), hsa-miR-103 (SEQ. ID NO: 3), hsa-miR- 107 (SEQ. ID NO: 5), hsa-miR-181a (SEQ. ID NO: 7), and hsa-miR-210 (SEQ. ID NO: 9) in as little as 0.25ρg of total RNA.
Interestingly, as indicated in Figure 4, the EBV-miRNAs BART3 (SEQ. ID NO: 33) and BHRF1-2 (SEQ. ID NO: 35) could be detected in as little as 0.25 pg of total cDNA (Fig. 5B). The less abundant miRNAs, processed from the other arm of their pre-miRNAs, the BART3* (SEQ. ID NO: 34) and the BHRF1-2* (SEQ. ID NO: 36) were detected as well, albeit at a higher concentration of cDNA (2.5 pg vs. 0.25 pg).
Assuming that the amount of total RNA is about 7 pg/cell, the General (antisense) Probe Detection assay could detect miRNAs, which are not highly abundant, from a single cell.
Example 2.4 General Probe Detection Assay detection of in vivo expression of EBV- miRNAs
Expression of EBV-miRNAs in vivo was tested using RNA extracted from Peripheral blood mononuclear cells (PBMC) isolated from 10 ml of whole blood with FicoU-Hypaque solution (GE Healthcare Biosciences, Uppsala, Sweden). The details of sequences are presented in table 4.
Table 4: Sequences and primers used in detection of in vivo expression of EBV-miRNAs
A specific primer for U6 (SEQ. ID NO: 25) was used for normalization.
The PBMCs were collected from three individuals: one of whom had an acute EBV infection [infectious mononucleosis (IM, with the following serological markers: positive for EBV-VCA IgG and IgM but negative for EBV-EBNA antibodies)], and two healthy carriers of EBV (positive for IgG antibodies to EBV-EBNA and EBV-VCA and negative for IgM VCA, indicating past and latent EBV infection). Since the expected number of B- cells that harbor EBV in PBMCs of healthy donors is estimated to be one out of 106 cells, and RNA was extracted from 107 PBMCs, the qPCR cycling of the General Probe Detection
Assay was extended to 75 cycles instead of the usual 40 cycles that were used otherwise. The assay showed a very high sensitivity; it was able to detect EBV-miRNAs in PBMCs of both the patient and the healthy carriers of EBV. As indicated in Fig. 6, a differential expression of various EBV-miRNAs was observed during the acute IM vs. the latent infection (healthy donors): BART2 (SEQ. ID NO: 32), BART4 (SEQ. ID NO: 37), and BART6 (SEQ. ID NO: 38) were clearly expressed in healthy carriers only, while BART7 (SEQ. ID NO: 39) was expressed only in the acute M. Regardless of the 75 cycles used in the qPCR, the "no-template-control" remained negative.
Example 3: Detection of cre-miR1142 (SEQ. ID NO: 51) by the General Probe Detection Assay and by SYBR Green
The Chlamydomonas reinhardtii algae, UTEX 90 strain (University of Texas algae collection, Austin TX) were maintained in TAP medium in an Adaptis growth chamber
(Conviron, Canada) at 22°C under constant white light at an intensity of 175 μmol photon m"2 s-1 while being shaken at 120 rpm in 250-ml flasks containing 50 ml of growth medium.
Total RNA was extracted using the mirVana™ kit (Ambion, Austin TX) following the instruction manual for total RNA isolation protocol using the following adaptations: 50ml of alga cells were harvested by centrifugation at 4,000 rpm for 10 minutes, washed once with 25ml of cold algae culture broth medium and re-suspended in 2ml of the mirVana™ kit Lysis/Binding Buffer. Cells were ground using a Polytron PT - MR 2100 homogenizer (Kinematica AG, Switzerland).
The extracted material was treated by DNase (Ambion), extracted by phenol and participated by ETOH.
Cre-mirR1142 (SEQ. ID NO: 51) was detected by both of the SYBR Green method and the General (antisense) Probe Detection Assay. The General probe used in this example
(SEQ. ID NO: 48) is an MGB probe which is partially complementary to the adaptor sequence region of the sense strand of the amplicon and further partially complementary to the poly (T) region of the sense strand of the amplicon.
Prior to the detection, the total RNA was incubated in the presence of a poly A polymerase enzyme (Takara, Otsu Japan), MnCl2, and ATP for 1 h at 37°C. Then, using an oligo dT tail harboring a consensus sequence, reverse transcription was performed on total
RNA using Superscript II RT (Invitrogen, Carlsbad CA). Next, the cDNA was amplified by real time PCR; this reaction contained a microRNA-specific forward primer and a universal
reverse primer complementary to the consensus 3' sequence of the oligo dT tail. The sequences used in the PCR procedure are detailed in table 5 below.
TABLE 5: Sequences used for amplification of Cre-mirR1142 (SEQ. ID NO: 51)
The protocol of the SYBR Green method and the General Probe Detection Assay is described in examples 3.1 and 3.2 below.
Example 3.1 Detection of cre-miRl 142 using the SYBR Green Method
1. Divide the Fwd primer (SEQ. ID NO: 52) into 96-well plate and spin it down.
2. Dilute the cDNA: 25ng/μl → 5ng/μl → 0.5ng/μl.
3. Assemble (at room temp .) :
4. Flip the tube, vortex and short spin. Put the mix on ice and add:
5. Divide 18 μl from the mix into each well. 6. Add Iμl Fwd primer (SEQ. ID NO: 52) and lμl DDW.
7. Short spin the plate and start the reaction.
8. The reaction program: stage 1, Reps = 1 step 1: Hold @ 95.0 C for 10:00 (MM:SS), Ramp Rate = 100 Stage 2, Reps = 42
Step 1: Hold @ 95.0 C for 0:15 (MM:SS)5 Ramp Rate = 100
Step 2: Hold @ 60.0 C for 1:00 (MM:SS), Ramp Rate = 100
Dissociation Protocol:
Stage 3, Reps = 1
Step 1: Hold @ 95.0 C for 0:15 (MM:SS), Ramp Rate = Auto
Step 2: Hold @ 60.0 C for 1 :00 (MM:SS), Ramp Rate = Auto
Step 3: Hold @ 95.0 C for 0:15 (MMrSS), Ramp Rate = Auto
Standard 7500 Mode
Sample Volume (μL): 20.0
Data Collection: Stage 2, Step 2
Example 3.2 Detection of cre-miR1142 using the General (antisense) Probe Detection
Assay
Centrifuge the Fwd primer set and the TaqMan probe (SEQ. ID NO: 48). 1. Take out the cDNA, defrost and keep on ice. Dilute the cDNA: 50ng/μl -» 5ng/μl →0.5ng/μl
2. Add in 2ml tube:
3. Flip the tube, vortex and short spin. Put the mix on ice.
4. Add:
6. Add 1 μl Fwd primer (SEQ. ID NO: 52) and 1 μl DDW.
7. Short spin the plate and start the reaction.
8. The reaction program : Stage 1, Reps = 1 step 1 : Hold @ 95.0 C for 10:00 (MMrSS), Ramp Rate = 100
Stage 2, Reps = 42
Step 1: Hold @ 95.0 C for 0:15 (MMrSS), Ramp Rate = 100 Step 2: Hold @ 60.0 C for 1:00 (MMrSS), Ramp Rate = 100
Standard 7500 Mode Sample Volume (μL): 20.0 Data Collection: Stage 2, Step 2
Example 3.3: Detection of cre-miRl 142 (SEQ. E) NO: 51) by the General Probe Detection Assay and by SYBR Green
The products of the qRT-PCR by each of the General Probe Detection Assay and SYBR Green (with water as a control) were run on agarose gel. As indicated in figures 7A and 7B, the same products were obtained in both detection methods, including primer- dimers and other unspecific products. Contrastingly, the Ct values were higher for the General Probe Detection Assay (32.96 Ct) than for the SYBR green method (29.87 Ct). The relatively high signal demonstrates specific detection of the cre-miRl 142 (SEQ. ID NO: 51) sequence, whereas the lower signal of SYBR Green is the result of dying unspecific product and primer-dimers, along with the cre-miRl 142 (SEQ. ID NO: 51) sequence.
Example 4; miR detection: with a specific probe, by the General (antisense) Probe Detection Assay and by the General (antisense) Probe Detection Assay with a forward primer harboring an elongated tail sequence
Example 4.1 Detection of target miR in tissue samples RNA material: Five samples of stomach tissue were obtained; FFPE samples were incubated repeatedly in xylene at 570C to remove excess paraffin, followed by washing in ethanol. Proteins were digested by proteinase K solution at 450C for a few hours. The RNA was extracted with acid phenol: chloroform, followed by ethanol precipitation and DNAse digestion. cDNA was prepared from the RNA. Each cDNA was diluted to 5 ng/μl and than to
0.5 ng/ μ\. The target nucleic acid, hsa-miR-451 (most abundant variant: SEQ. ID NO: 54) was detected in the cDNA by the three following methods:
1. With a forward primer (SEQ. ID NO: 55) and a specific probe (SEQ. ID NO: 56) [and with U6 (SEQ. ID NO: 60) to normalize the results] 2. With a forward primer (SEQ. ID NO: 57), and the General antisense MGB Probe
(SEQ. ID NO: 48) which is partially complementary to the adaptor sequence region of the sense strand and to the poly(T) region of the sense strand of the amplicon.
3. With a forward primer with an elongated tail (SEQ. ID NO: 58) and the General antisense Probe (SEQ. ID NO: 48) as above.
The same reverse primer (SEQ. ID NO: 28) was used in all of the qPCR reactions.
It should be noted that the General Probe Assay (sense and antisense) allows for detection of multiple variants of the targeted miR, as opposed to the assay with the specific probe, which detects only a specific variant. The sequences used in the detection are detailed in table 6.
TABLE 6: Sequences used in the detection of hsa-miR-451 (SEQ. ID NO: 54)
The qPCR reaction included 42 cycles.
The results of the detection for one exemplary sample are detailed in tables 7.1 and
7.2.
Table 7.1: Detection of hsa-miR-451 (SEQ. ID NO: 54) with a specific probe (in Cts, average of triplicates)
Table 7.2: detection of hsa-miR-451 (SEQ. ID NO: 54) (in Cts, average of triplicates) with a General antisense probe
As apparent from tables 7.1 and 7.2, the average Ct, in both dilutions, is lower for the detection with the General probe than for the detection with the specific probe. This result implies that the hybridization of the forward primer used in the General (antisense) Probe Detection Assay to the target nucleic acid is improved as compared to the hybridization of the forward primer used in the detection with the specific probe, thereby enhancing the effectiveness of the detection. Moreover, the average Ct (in both dilutions) is lowest for the detection with the General probe using a forward primer with an elongated tail. The elongation of the tail raises the TM of the forward primer to 6O0C at least.
Example 4.2 Detection of synthetic hsa-miR-451 (SEP. ID NO: 59)
Synthetic hsa-miR-451 (SEQ. ID NO: 59) was used to prepare 2.5nM cDNA. Synthetic hsa-miR-451 (SEQ. ID NO: 59) was detected with a specific probe (SEQ. ID NO: 56) and a forward primer (SEQ. ID NO: 55), and also with the General antisense MGB Probe (SEQ. ID NO: 48, which is partially complementary to the adaptor sequence region of the sense strand and to the poly(T) region of the sense strand of the amplicon) and a forward primer with an elongated tail (SEQ. ID NO: 58). The same reverse primer (SEQ. ID NO: 28) was used in both qPCR reactions.
The sequences used in the detection are detailed in table 8 below. TABLE 8: Sequences used in the detection of synthetic hsa-miR-451 (SEQ. ID NO: 59)
Sequence (5'-3') SEQ ID NO:
Synthetic hsa-miR-451 AAACCGUUACCAUUACUGAGUU 59 forward primer (for assay
CAGTCATTTGGGAAACCGTTACCATTAC ' 55 with specific probe)
Specific hsa-miR-451probe TCCGTTTTTTTTTTTTACTCAGTA 56 forward primer with elongated tail (for assay CATTTGGAAACCGTTACCATTACTGAGT 58 with General probe)
General antisense probe CGACTCACTATCGGTTTT 48
Reverse primer GCGAGCACAGAATTAATACGAC 28
The RT-PCR reaction included 55 cycles. The results of the detection are detailed in table 9.
Table 9: Detection of synthetic hsa-miR-451 (SEQ. ID NO: 59) (in Cts, average of duplicates) by the General (antisense) Probe Detection Assay, and by use of a specific probe
As shown in table 9, the average Ct, in all of the dilutions, is lower for the detection with the General antisense Probe than for the detection with the specific probe. This finding is in accordance with the results obtained in the detection of hsa-miR-451 (SEQ. ID NO: 54) in tissue samples as described in Example 4.1 above; this negates the possibility that the low Ct's of the General (antisense) Probe detection Assay are the result of non-specific detection of nucleic acids, since no other nucleic acids besides the synthetic hsa-miR-451 (SEQ. ID NO: 59) were in the tested samples.
Example 5: General Probe Detection Assay: detection with a General sense Probe According to some embodiments, the probe of the General Probe Detection Assay is an MGB probe which is complementary to the adaptor sequence region and to the poly(T) region of the antisense strand of the amplicon. This probe is designated General sense Probe, and it may be reverse complement to the General antisense probe described in examples 3 and 4 above. Example 5.1 Sensitivity of General (sense) Probe Detection Assay
The sensitivity of the assay and the ability to detect specific miRNAs in the presence of other miRNAs were determined in a competition assay, as follows:
Total RNA from fresh liver samples and from brain samples was obtained from Ambion. For each of the tissues cDNA was prepared from the RNA, and serially diluted in the cDNA of the other tissue to a final concentration of 0.5 ng/μg liver-brain cDNA mixture.
The General (sense) Probe Detection Assay was then used to detect each of the liver- specific hsa-miR-122 (SEQ. ID NO: 61) and the brain-specific hsa-miR-124 (SEQ. ID NO: 62) in the mixtures.
The sequences used in the detection are detailed in table 10 below.
Table 10: Sequences used in detection of hsa-miR-122 (SEQ. ID NO: 61) and hsa-miR- 124 (SEQ. ID NO: 62) by General (sense) Probe Detection Assay
SEQ
Sequence (5'-3') ID NO:
Forward primer hsa-miR-122 GGCTGGAGTGTGACAATGGTGTTT 63
Forward primer hsa-miR- 124 TTAAGGCACGCGGTGAATGCC 64
General sense probe AAAACCGATAGTGAGTCG 49
Reverse primer GCGAGCACAGAATTAATACGAC 28
The results are detailed in tables 11.1 and 11.2 and in figures 8 A and 8B.
Table 11.1: Detection of liver-specific hsa-miR-122 (SEQ. ID NO: 61) spiked in cDNA of Brain tissues
Table 11.2: Detection of brain-specific hsa-miR-124 (SEQ. ID NO: 62) spiked in cDNA of Liver tissues
The results in tables 11.1 and 11.2 and in figures 8 A and 8B demonstrate the wide dynamic range of the General (sense) Probe Detection Assay, which was able to detect the target miR in concentrations spanning 16 Cts (216 fold) from 100% to 0% liver tissue and 12 Cts (212 fold) from 100% to 0% brain tissue.
The results further demonstrate the high sensitivity of the General (sense) Probe Detection Assay, which detected as little as 0.00152588% of the liver-specific hsa-miR-122
(SEQ. ID NO: 61), and 0.048828125% of the brain-specific hsa-miR-124 (SEQ. ID NO: 62), in the mixture of cDNA of total RNA from Liver and Brain tissues.
This sensitivity of the General Probe Detection Assay may be highly advantageous, for example, in the detection of minute concentrations of tumor tissue metastases to other tissue or organ.
Example 5.2 Specificity of General (sense) Probe Detection Assay
The ability of the General (sense) Probe Detection Assay to discriminate between miRNAs within a family was tested on the let-7 family. cDNA libraries were prepared from synthetic miRNA sequences of hsa-let-7a, hsa-let-7b, hsa-let-7c and hsa-let-7d (SEQ. ID NO: 65-68 respectively). Each of these sequences were then amplified using the General (sense) Probe Detection Assay, with sequences of hsa-let-7a to -7d as forward-primer- detectors, as detailed in table 12 below.
Table 12: Sequences used in the cross-reactivity detection assay
Forward primers (5 '-3') SEQ. ID NO hsa-let-7a TCATTTGGCTGAGGTAGTAGGTTGTATAGTT 69 hsa-let-7b CATTTGGTGAGGTAGTAGGTTGTGTGGTT 70 hsa-let-7c CATTTGGTGAGGTAGTAGGTTGTATGGTT 71 hsa-let-7d CATTTGGAGAGGTAGTAGGTTGCATAGT 72
General sense AAAACCGATAGTGAGTCG 49 probe
Reverse primer GCGAGCACAGAATTAATACGAC 28
The results of the detection for one exemplary sample are detailed in table 13. Table 13: Cross reactivity (in Cts) between members of the hsa-let-7 family, as detected with the sense probe of the General Probe Detection Assay
The cross-reactivity experiment highlights the specificity of the General' (sense)
Probe Detection Assay. The results in table 13 demonstrate specific detection of synthetic hsa-let-7a (SEQ. ID NO: 65), synthetic hsa-let-7b (SEQ. ID NO: 66) and synthetic hsa-let- 7d (SEQ. ID NO: 68) by their respective corresponding forward primers (lowest Cts values), while much lower signals (i.e., higher Ct values) are observed for the non-specific amplification of very similar sequences. This specificity is further demonstrated by the ability of the General Probe Detection Assay, using a specific forward primer and the
General sense MGB Probe, to detect each of the specific synthetic sequences in a mixture with other family members. This is essential for detection of specific biomarker-miRs in tissues comprising multiple miRs.
Example 6: General Probe Detection Assay: detection in serum
The discovery of circulating microRNA in human serum has opened up new possibilities for non-invasive diagnosis. The detection of specific miRs in serum by the General Probe Detection Assay was tested as described below.
RNA extraction: 100 ml serum was incubated at 560C for 1 h with 0.65 mg/ml Proteinase K (Sigma P2308). Two synthetic RNAs (IDT) were spiked-in as controls before acid phenolxhloroform extraction and then RNA was ETOH precipitated overnight at - 200C.
Next, DNase treatment was performed to eliminate residual DNA fragments. Finally, after a second acid phenolxhloroform extraction, the pellet was re-suspended in DDW and two additional synthetic RNAs were spiked-in as controls. Each of the miRs listed in table 14 was specifically detected in the serum samples by the General Probe detection Assay, with both of the antisense (SEQ. ID NO: 48) and sense (SEQ. ID NO: 49) MGB probes.
Table 14: Detection of miRs in serum (in Cts), with General Probe Detection Assay
The foregoing description of the specific embodiments will so fully reveal the general nature of the invention that others can, by applying current knowledge, readily modify and/or adapt for various applications such specific embodiments without undue experimentation and without departing from the generic concept, and, therefore, such adaptations and modifications should and are intended to be comprehended within the meaning and range of equivalents of the disclosed embodiments. Although the invention has been described in conjunction with specific embodiments thereof, it is evident that many alternatives, modifications and variations will be apparent to those skilled in the art.
Accordingly, it is intended to embrace all such alternatives, modifications and variations that fall within the spirit and broad scope of the appended claims.
It should be understood that the detailed description and specific examples, while indicating preferred embodiments of the invention, are given by way of illustration only, since various changes and modifications within the spirit and scope of the invention will become apparent to those skilled in the art from this detailed description.
Claims
1. A method of detecting a target RNA nucleic acid sequence in a biological sample, the method comprising: a) providing the biological sample comprising the target RNA nucleic acid sequence; b) annealing the target RNA nucleic acid sequence with a poly(T) primer comprising a 5' universal adaptor sequence; c) generating a reverse transcript of the polyadenylated RNA with the poly(T) primer of step (b); and d) amplifying the reverse transcript of step (c) by a polymerase chain reaction (PCR) comprising a specific forward primer, a universal reverse primer and a general probe, to generate an amplicon; wherein the forward primer is at least partially identical to the target RNA nucleic acid sequence and the universal reverse primer is at least partially identical to a 5' region of the adaptor sequence of the poly (T) primer.
2. The method of claim 1, wherein the target RNA nucleic acid sequence is a short nucleic acid, and wherein said target RNA sequence of step (a) is extended at the 31 end by polyadenylation prior to step (b).
3. The method of claim 1, wherein the general probe is partially complementary to the adaptor sequence region of the sense strand of the amplicon.
4. The method of claim 3, wherein the general probe is further partially complementary to the poly(T) region of the sense strand of the amplicon.
5. The method of claim 1, wherein the general probe is complementary to the adaptor sequence region of the antisense strand of the amplicon.
6. The method of claim 5, wherein the general probe is further partially complementary to the poly(T) region of the antisense strand of the amplicon.
7. The method of claim 3, wherein the general probe comprises a sequence selected from the group consisting of SEQ. ID NOS: 48 and 50.
8. The method of claim 4, wherein the general probe comprises SEQ. ID. NO: 48.
9. The method of claim 6, wherein the general probe comprises SEQ. ID. NO: 49.
10. The method of claim 2, wherein the target RNA nucleic acid sequence is selected from the group consisting of: microRNA, siRNA, PiwiRNA, and a combination thereof.
11. The method of claim 2, wherein the target RNA nucleic acid sequence is 17-25 nt in length.
12. The method of claim 1, wherein the general probe is a TaqMan probe.
13. The method of claim 1, wherein the general probe comprises a fluorescent reporter group and a quencher.
14. The method of claim 13, wherein said general probe comprises a 5' fluorescent reporter group and 3' a quencher.
15. The method of claim 1, wherein said general probe further comprises a minor groove binder (MGB).
16. The method of claim 1, wherein said general probe further comprises locked nucleic acids (LNA).
17. The method of claim 1, wherein the forward primer comprises a sequence selected from the group consisting of SEQ. ID NOS. 2, 4, 6, 8, 12, 14, 16, 18, 20, 22, 24, 40- 47, 52, 55, 57, 58, 63, 64, 69-87, 89 and 91.
18. The method of claim 1, wherein the reverse primer comprises the sequence of SEQ. ID NO. 28.
19. The method of claim 1, wherein said biological sample is selected from the group consisting of bodily fluid, a cell line, a tissue sample, a biopsy sample, a needle biopsy sample, a surgically removed sample, and a sample obtained by tissue- sampling procedures.
20. The method of claim 19, wherein said bodily fluid is serum.
21. The method of claim 19, wherein said tissue is a fresh, frozen, fixed, wax-embedded or formalin-fixed paraffin-embedded (FFPE) tissue.
22. The method of claim 1, wherein said biological sample is derived from a plant.
23. A kit for the detection of a target RNA nucleic acid sequence in a biological sample, the kit comprising a poly(T) primer comprising a 5' universal adaptor sequence, a specific forward primer, a universal reverse primer and a general probe, wherein the forward primer is at least partially identical to the target RNA nucleic acid sequence, and the universal reverse primer is at least partially identical to a 5' region of the adaptor sequence of the poly (T) primer.
24. The kit of claim 23, wherein the general probe comprises a sequence selected from the group consisting of SEQ. ID NOS: 48-50.
25. The kit of claim 23, wherein the target RNA nucleic acid sequence is a short RNA nucleic acid sequence selected from the group consisting of: microRNA, siRNA, PiwiRNA, and a combination thereof.
26. The kit of claim 25, wherein the target RNA nucleic acid sequence is 17-25 nt in length.
27. The kit of claim 23, wherein the general probe is a TaqMan probe.
28. The kit of claim 23, wherein the general probe comprises a fluorescent reporter group and a quencher.
29. The kit of claim 28, wherein said general probe further comprises a minor groove binder (MGB).
30. The kit of claim 23, wherein said general probe further comprises locked nucleic acids (LNA).
31. The kit of claim 23, wherein the forward primer comprises a sequence selected from the group consisting of SEQ. ID NOS. 2, 4, 6, 8, 12, 14, 16, 18, 20, 22, 24, 40-47, 52, 55, 57, 58, 63, 64, 69-87, 89 and 91.
32. The kit of claim 23, wherein the reverse primer comprises the sequence of SEQ. ID NO. 28.
Applications Claiming Priority (2)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| US15877609P | 2009-03-10 | 2009-03-10 | |
| US61/158,776 | 2009-03-10 |
Publications (1)
| Publication Number | Publication Date |
|---|---|
| WO2010103522A1 true WO2010103522A1 (en) | 2010-09-16 |
Family
ID=42727869
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| PCT/IL2010/000206 WO2010103522A1 (en) | 2009-03-10 | 2010-03-10 | Method for detection of nucleic acid sequences |
Country Status (1)
| Country | Link |
|---|---|
| WO (1) | WO2010103522A1 (en) |
Cited By (4)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| EP2700719A1 (en) * | 2011-04-20 | 2014-02-26 | Shenzhen University | METHOD AND PRIMERS FOR DETECTION OF miRNA, AND APPLICATION THEREOF |
| WO2017164353A1 (en) * | 2016-03-25 | 2017-09-28 | 参天製薬株式会社 | Detection method and detection kit for detecting dry age-related macular degeneration |
| WO2018165593A1 (en) * | 2017-03-09 | 2018-09-13 | iRepertoire, Inc. | Dimer avoided multiplex polymerase chain reaction for amplification of multiple targets |
| CN113373203A (en) * | 2021-08-16 | 2021-09-10 | 北京恩泽康泰生物科技有限公司 | Design method and application of primer and probe combination for detecting miRNA |
Citations (9)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US20060051771A1 (en) * | 2004-09-07 | 2006-03-09 | Ambion, Inc. | Methods and compositions for tailing and amplifying RNA |
| US20060094025A1 (en) * | 2004-11-02 | 2006-05-04 | Getts Robert C | Methods for detection of microrna molecules |
| WO2006126040A1 (en) * | 2005-05-25 | 2006-11-30 | Rosetta Genomics Ltd. | Bacterial and bacterial associated mirnas and uses thereof |
| US20070003575A1 (en) * | 2004-05-26 | 2007-01-04 | Itzhak Bentwich | Viral and viral associated MiRNAs and uses thereof |
| US7232806B2 (en) * | 2001-09-28 | 2007-06-19 | Max-Planck-Gesellschaft Zur Foerderung Der Wissenschaften E.V. | MicroRNA molecules |
| WO2007074405A2 (en) * | 2005-09-16 | 2007-07-05 | Devgen Nv | Dsrna as insect control agent |
| US20080045418A1 (en) * | 2005-02-04 | 2008-02-21 | Xia Xueliang J | Method of labeling and profiling rnas |
| WO2008104985A2 (en) * | 2007-03-01 | 2008-09-04 | Rosetta Genomics Ltd. | Methods for distingushing between lung squamous carcinoma and other non smallcell lung cancers |
| US20100047784A1 (en) * | 2006-06-30 | 2010-02-25 | Rosetta Genomics Ltd. | Method for detecting nucleic acids |
-
2010
- 2010-03-10 WO PCT/IL2010/000206 patent/WO2010103522A1/en active Application Filing
Patent Citations (9)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| US7232806B2 (en) * | 2001-09-28 | 2007-06-19 | Max-Planck-Gesellschaft Zur Foerderung Der Wissenschaften E.V. | MicroRNA molecules |
| US20070003575A1 (en) * | 2004-05-26 | 2007-01-04 | Itzhak Bentwich | Viral and viral associated MiRNAs and uses thereof |
| US20060051771A1 (en) * | 2004-09-07 | 2006-03-09 | Ambion, Inc. | Methods and compositions for tailing and amplifying RNA |
| US20060094025A1 (en) * | 2004-11-02 | 2006-05-04 | Getts Robert C | Methods for detection of microrna molecules |
| US20080045418A1 (en) * | 2005-02-04 | 2008-02-21 | Xia Xueliang J | Method of labeling and profiling rnas |
| WO2006126040A1 (en) * | 2005-05-25 | 2006-11-30 | Rosetta Genomics Ltd. | Bacterial and bacterial associated mirnas and uses thereof |
| WO2007074405A2 (en) * | 2005-09-16 | 2007-07-05 | Devgen Nv | Dsrna as insect control agent |
| US20100047784A1 (en) * | 2006-06-30 | 2010-02-25 | Rosetta Genomics Ltd. | Method for detecting nucleic acids |
| WO2008104985A2 (en) * | 2007-03-01 | 2008-09-04 | Rosetta Genomics Ltd. | Methods for distingushing between lung squamous carcinoma and other non smallcell lung cancers |
Cited By (11)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| EP2700719A1 (en) * | 2011-04-20 | 2014-02-26 | Shenzhen University | METHOD AND PRIMERS FOR DETECTION OF miRNA, AND APPLICATION THEREOF |
| EP2700719A4 (en) * | 2011-04-20 | 2014-12-03 | Univ Shenzhen | METHOD AND PRIMERS FOR DETECTION OF miRNA, AND APPLICATION THEREOF |
| WO2017164353A1 (en) * | 2016-03-25 | 2017-09-28 | 参天製薬株式会社 | Detection method and detection kit for detecting dry age-related macular degeneration |
| JPWO2017164353A1 (en) * | 2016-03-25 | 2019-02-07 | 参天製薬株式会社 | Test kit and test method for atrophic age-related macular degeneration |
| WO2018165593A1 (en) * | 2017-03-09 | 2018-09-13 | iRepertoire, Inc. | Dimer avoided multiplex polymerase chain reaction for amplification of multiple targets |
| CN110662756A (en) * | 2017-03-09 | 2020-01-07 | 艾瑞普特公司 | Multiplex polymerase chain reaction avoiding dimers for multi-target amplification |
| JP2020509747A (en) * | 2017-03-09 | 2020-04-02 | アイレパートリー インコーポレイテッド | Dimer avoidance multiplex polymerase chain reaction for amplifying multiple targets |
| US20220259653A1 (en) * | 2017-03-09 | 2022-08-18 | iRepertoire, Inc. | Dimer avoided multiplex polymerase chain reaction for amplification of multiple targets |
| JP7280191B2 (en) | 2017-03-09 | 2023-05-23 | アイレパートリー インコーポレイテッド | Dimer Avoidance Multiplex Polymerase Chain Reaction to Amplify Multiple Targets |
| CN110662756B (en) * | 2017-03-09 | 2023-09-15 | 艾瑞普特公司 | Dimer-avoiding multiplex polymerase chain reaction for multi-target amplification |
| CN113373203A (en) * | 2021-08-16 | 2021-09-10 | 北京恩泽康泰生物科技有限公司 | Design method and application of primer and probe combination for detecting miRNA |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| JP5851496B2 (en) | Modified stem-loop oligonucleotide-mediated reverse transcription and quantitative PCR with limited base spacing | |
| US8314220B2 (en) | Methods compositions, and kits for detection of microRNA | |
| US9169520B2 (en) | Cancer marker, method for evaluation of cancer by using the cancer marker, and evaluation reagent | |
| US7993831B2 (en) | Methods of normalization in microRNA detection assays | |
| US9096895B2 (en) | Method for quantification of small RNA species | |
| CN113604564A (en) | Method for detecting exosome-associated microRNA (ribonucleic acid) molecules | |
| EP2900836B1 (en) | Two-primer pcr for microrna multiplex assay | |
| WO2016186987A1 (en) | Biomarker micrornas and method for determining tumor burden | |
| US20160177376A1 (en) | Detection method of micro-rna with high specificity | |
| Lan et al. | Linear-hairpin variable primer RT-qPCR for MicroRNA | |
| CN101082060A (en) | New micro ribonucleic acid quantitative PCR (polymerase chain reaction) detection method | |
| JP2015512249A (en) | Method for determining affinity and receptor usage of viruses, especially HIV, in body samples taken from the circulation | |
| EP2691545A2 (en) | Methods for lung cancer classification | |
| JP2019524103A (en) | RNA detection kit and method | |
| CA2677043A1 (en) | Method for distinguishing between lung squamous carcinoma and other non small cell lung cancers | |
| WO2010103522A1 (en) | Method for detection of nucleic acid sequences | |
| CN107385037B (en) | MiRNA indirect real-time fluorescence quantitative PCR detection method | |
| Castoldi et al. | 22 Expression Profiling of MicroRNAs by Quantitative Real-Time PCR | |
| WO2010058393A2 (en) | Compositions and methods for the prognosis of colon cancer | |
| US8563252B2 (en) | Methods for distinguishing between lung squamous carcinoma and other non small cell lung cancers | |
| WO2012149222A2 (en) | Methylated coding and non-coding rna genes as diagnostic and therapeutic tools for human melanoma | |
| Castoldi et al. | 22 Expression Profiling of | |
| Wang et al. | RT2 miRNA PCR Arrays | |
| Jonstrup | Discovering miRNAs: Ph. D. thesis |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| 121 | Ep: the epo has been informed by wipo that ep was designated in this application |
Ref document number: 10750456 Country of ref document: EP Kind code of ref document: A1 |
|
| NENP | Non-entry into the national phase |
Ref country code: DE |
|
| 122 | Ep: pct application non-entry in european phase |
Ref document number: 10750456 Country of ref document: EP Kind code of ref document: A1 |