WO2004108964A1 - Differentially regulated hepatocellular carcinoma genes and uses thereof - Google Patents
Differentially regulated hepatocellular carcinoma genes and uses thereof Download PDFInfo
- Publication number
- WO2004108964A1 WO2004108964A1 PCT/SG2004/000166 SG2004000166W WO2004108964A1 WO 2004108964 A1 WO2004108964 A1 WO 2004108964A1 SG 2004000166 W SG2004000166 W SG 2004000166W WO 2004108964 A1 WO2004108964 A1 WO 2004108964A1
- Authority
- WO
- WIPO (PCT)
- Prior art keywords
- nucleic acid
- acid molecules
- expression
- sample
- hcc
- Prior art date
Links
Classifications
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12N—MICROORGANISMS OR ENZYMES; COMPOSITIONS THEREOF; PROPAGATING, PRESERVING, OR MAINTAINING MICROORGANISMS; MUTATION OR GENETIC ENGINEERING; CULTURE MEDIA
- C12N9/00—Enzymes; Proenzymes; Compositions thereof; Processes for preparing, activating, inhibiting, separating or purifying enzymes
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P1/00—Drugs for disorders of the alimentary tract or the digestive system
- A61P1/16—Drugs for disorders of the alimentary tract or the digestive system for liver or gallbladder disorders, e.g. hepatoprotective agents, cholagogues, litholytics
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P35/00—Antineoplastic agents
-
- A—HUMAN NECESSITIES
- A61—MEDICAL OR VETERINARY SCIENCE; HYGIENE
- A61P—SPECIFIC THERAPEUTIC ACTIVITY OF CHEMICAL COMPOUNDS OR MEDICINAL PREPARATIONS
- A61P43/00—Drugs for specific purposes, not provided for in groups A61P1/00-A61P41/00
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K14/00—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof
- C07K14/435—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof from animals; from humans
- C07K14/46—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof from animals; from humans from vertebrates
- C07K14/47—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof from animals; from humans from vertebrates from mammals
-
- C—CHEMISTRY; METALLURGY
- C07—ORGANIC CHEMISTRY
- C07K—PEPTIDES
- C07K14/00—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof
- C07K14/435—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof from animals; from humans
- C07K14/46—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof from animals; from humans from vertebrates
- C07K14/47—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof from animals; from humans from vertebrates from mammals
- C07K14/4701—Peptides having more than 20 amino acids; Gastrins; Somatostatins; Melanotropins; Derivatives thereof from animals; from humans from vertebrates from mammals not used
- C07K14/4748—Tumour specific antigens; Tumour rejection antigen precursors [TRAP], e.g. MAGE
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12Q—MEASURING OR TESTING PROCESSES INVOLVING ENZYMES, NUCLEIC ACIDS OR MICROORGANISMS; COMPOSITIONS OR TEST PAPERS THEREFOR; PROCESSES OF PREPARING SUCH COMPOSITIONS; CONDITION-RESPONSIVE CONTROL IN MICROBIOLOGICAL OR ENZYMOLOGICAL PROCESSES
- C12Q1/00—Measuring or testing processes involving enzymes, nucleic acids or microorganisms; Compositions therefor; Processes of preparing such compositions
- C12Q1/68—Measuring or testing processes involving enzymes, nucleic acids or microorganisms; Compositions therefor; Processes of preparing such compositions involving nucleic acids
- C12Q1/6876—Nucleic acid products used in the analysis of nucleic acids, e.g. primers or probes
- C12Q1/6883—Nucleic acid products used in the analysis of nucleic acids, e.g. primers or probes for diseases caused by alterations of genetic material
- C12Q1/6886—Nucleic acid products used in the analysis of nucleic acids, e.g. primers or probes for diseases caused by alterations of genetic material for cancer
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12Q—MEASURING OR TESTING PROCESSES INVOLVING ENZYMES, NUCLEIC ACIDS OR MICROORGANISMS; COMPOSITIONS OR TEST PAPERS THEREFOR; PROCESSES OF PREPARING SUCH COMPOSITIONS; CONDITION-RESPONSIVE CONTROL IN MICROBIOLOGICAL OR ENZYMOLOGICAL PROCESSES
- C12Q2600/00—Oligonucleotides characterized by their use
- C12Q2600/106—Pharmacogenomics, i.e. genetic variability in individual responses to drugs and drug metabolism
-
- C—CHEMISTRY; METALLURGY
- C12—BIOCHEMISTRY; BEER; SPIRITS; WINE; VINEGAR; MICROBIOLOGY; ENZYMOLOGY; MUTATION OR GENETIC ENGINEERING
- C12Q—MEASURING OR TESTING PROCESSES INVOLVING ENZYMES, NUCLEIC ACIDS OR MICROORGANISMS; COMPOSITIONS OR TEST PAPERS THEREFOR; PROCESSES OF PREPARING SUCH COMPOSITIONS; CONDITION-RESPONSIVE CONTROL IN MICROBIOLOGICAL OR ENZYMOLOGICAL PROCESSES
- C12Q2600/00—Oligonucleotides characterized by their use
- C12Q2600/136—Screening for pharmacological compounds
Definitions
- the invention is in the field of diagnostics and therapeutics for cancer. More specifically, the invention is in the field of diagnostics and therapeutics for hepatocellular carcinoma.
- Hepatocellular carcinoma is the most common primary malignant tumor of the liver that accounts for more than 70% of liver cancers worldwide (Parkin et al., 1999). Many risk factors have been associated with the development of HCC, including hepatitis B (HBV) and hepatitis C (HCV) viral infection, cirrhosis, male gender, exposure to toxins, etc. Death generally occurs due to liver failure associated with cirrhosis and/or rapid outgrowth of multiple nodules. Approximately 0.25-1 million new cases of HCC are diagnosed each year, and the cancer is especially prevalent in Southeast Asia, China, and sub-Saharan Africa. While surgical resection is considered to be the main curative treatment, only 10-15% of cases are suitable for surgery at the time of presentation. This is because either the disease is detected at an advanced stage at presentation or the underlying poor liver functional reserve precluded surgical intervention.
- HCC hepatitis B
- HCV hepatitis C
- HCC has included detection of the presence of a liver mass on radiological investigations and the detection of elevated serum alpha fetoprotein (AFP) levels (Yu and Keeffe, 2003).
- AFP serum alpha fetoprotein
- elevation of AFP is not exclusive to HCC and has been observed in benign hepatic disease, such as liver cirrhosis, and other cancers such as germ cell cancer (Bosl and Head, 1994).
- Treatment of HCC has included interferon therapy and antiviral drugs, but the results have proved unpredictable and the effectiveness maybe limited (Lee, 1997; Yu and Keeffe, 2003).
- Microarrays have been used to address changes in gene expression of HCC (Chen et al, 2002, Okabe et al, 2001; Nissan et al, 2001; Sbirota et al, 2001; Tackels-Horne et al, 2001; Xu et al, 2001a; Xu et al, 2001b).
- HCC Habal et al, 2002, Okabe et al, 2001; Nissan et al, 2001; Sbirota et al, 2001; Tackels-Horne et al, 2001; Xu et al, 2001a; Xu et al, 2001b.
- these reports were restricted to the tissue samples selected for each study and exhibited wide variation in the results, thus limiting the potential significance and utility of the data.
- the invention provides in part molecular markers for hepatocellular carcinoma (HCC) that may be used for HCC diagnosis, to assess HCC progression or regression, or the efficacy and/or toxicity of HCC therapeutics, and/or to identify candidate compounds for HCC therapy, with high predictive accuracy.
- HCC hepatocellular carcinoma
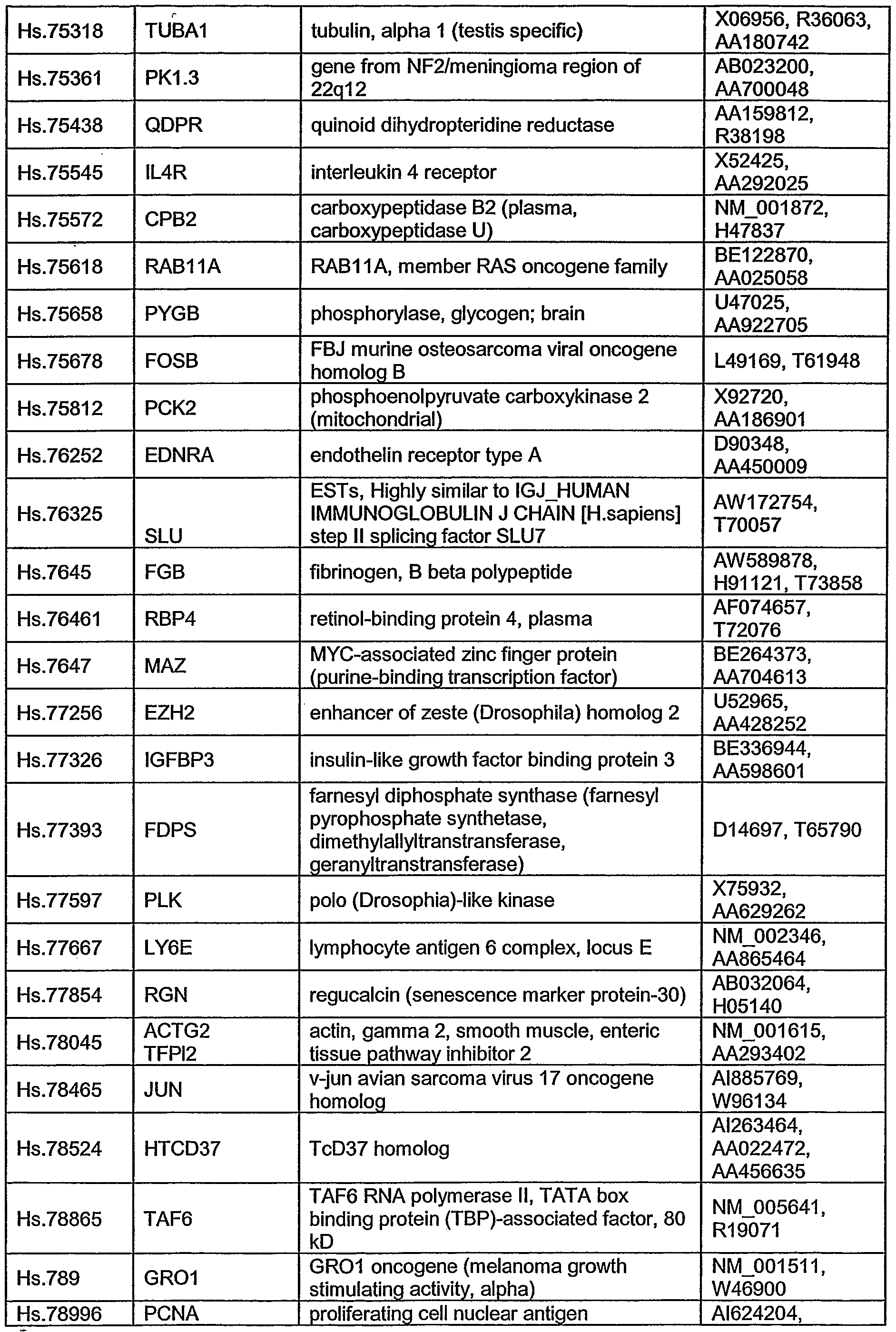
- the invention provides a composition including an addressable collection of two or more nucleic acid molecules, or polypeptides encoded by these nucleic acid molecules, that are differentially expressed in hepatocellular carcinoma, where the nucleic acid molecules consist essentially of the nucleic acid molecules set forth in any one or more of Tables 1 through 4 or complements, fragments, variants, or analogs thereof.
- the composition may include all of the nucleic acid molecules, or their encoded polypeptides, set forth in any one or more of Tables 1 through 4 or complements, fragments, variants, or analogs thereof, or any subset of these nucleic acid molecules or polypeptides.
- the nucleic acid molecules or polypeptides may be differentially expressed between hepatocellular carcinoma tissue and non-tumor tissue.
- the nucleic acid molecules or the polypeptides may be attached to a solid support.
- the compositions may be used in the preparation of a medicament for diagnosis or therapy of hepatocellular carcinoma.
- the invention provides a method of diagnosing hepatocellular carcinoma in a subject by obtaining a sample from the subject and detecting the level of expression of two or more nucleic acid molecules or expression products thereof in the sample, where the nucleic acid molecules consist essentially of the nucleic acid molecules set forth in any one or more of Tables 1 through 4 or complements, fragments, variants, or analogs thereof.
- the invention provides a method of monitoring the progression of hepatocellular carcinoma in a subject by obtaining a sample from the subject and detecting the level of expression of two or more nucleic acid molecules or expression products thereof in the sample, where the nucleic acid molecules consist essentially of the nucleic acid molecules set forth in any one or more of Tables 1 through 4, or complements, fragments, variants, or analogs thereof.
- the sample may be obtained at two or more time points.
- the method may further include comparing the level of expression of the nucleic acid molecules or expression products at two or more time points.
- the invention provides a method of monitoring the efficacy of a hepatocellular carcinoma therapy in a subject by administering the therapy to the subject, obtaining a sample from the subject, and detecting the level of expression of two or more nucleic acid molecules or expression products thereof in the sample, where the nucleic acid molecules consist essentially of the nucleic acid molecules set forth in any one or more of Tables 1 through 4, or complements, fragments, variants, or analogs thereof.
- the therapy maybe administered at two or more administration time points.
- the sample may be obtained at two or more sampling time points.
- the method may further include comparing the level of expression of the nucleic acid molecules or expression products at two or more administration time points, and/or at two or more sampling time points.
- the invention provides a method of screening a compound for treating hepatocellular carcinoma by contacting a sample with a test compound and detecting the level of expression of two or more nucleic acid molecules or expression products thereof in the sample, where the nucleic acid molecules consist essentially of the nucleic acid molecules set forth in any one or more of Tables 1 through 4, or complements, fragments, variants or analogs thereof.
- the sample may be liver or serum, or may be suspected of being cancerous, or may be non-cancerous.
- the methods may further include comparing the level of expression of the nucleic acid molecules or expression products thereof in a non-cancerous sample and in a sample suspected of being cancerous. Differential expression of the nucleic acid molecules or expression products thereof may be indicative of hepatocellular carcinoma, or of progression of hepatocellular carcinoma, or of the efficacy of the hepatocellular carcinoma therapy.
- the subject may be suspected of having hepatocellular carcinoma.
- the subject may be a human.
- the method may further include comparing the level of expression of two or more nucleic acid molecules or expression products thereof with a standard, or further include preparing a gene expression profile.
- the method may be a high throughput method.
- the invention provides a solid support including two or more nucleic acid molecules or polypeptides encoded by these nucleic acid molecules that are differentially expressed in hepatocellular carcinoma, where the nucleic acid molecules consist essentially of the nucleic acid molecules set forth in Tables 1 through 4 or complements, fragments, variants, or analogs thereof.
- the nucleic acid molecules may consist essentially of all the nucleic acid molecules set forth in any one or more of Tables 1 through 4, and/or be differentially expressed between hepatocellular carcinoma tissue and non-tumor tissue.
- the polypeptides may consist essentially of the polypeptides encoded by all the nucleic acid molecules set forth in any one or more of Tables 1 through 4, and/or be differentially expressed between hepatocellular carcinoma tissue and non-tumor tissue.
- the nucleic acid molecules or the polypeptides may be covalently or non-covalently attached to the solid support (e.g., a microarray).
- the invention provides a database including information identifying the expression level in liver tissue (e.g., cancerous or non-cancerous tissue) of two or more nucleic acid molecules or expression products thereof, where the nucleic acid molecules consist essentially of the nucleic acid molecules set forth in any one or more of Tables 1 through 4, or complements, fragments, variants, or analogs thereof.
- composition includes a plurality of the nucleic acid molecules described herein, including complements, analogs, variants, and fragments thereof.
- a composition as used herein also includes a plurality of polypeptides encoded by the nucleic acid molecules described herein, and complements, analogs, variants, and fragments thereof.
- a composition as used herein also includes a plurality of polypeptides capable of specifically binding to the polypeptides or nucleic acid molecules described herein (e.g., antibodies).
- the composition may include any combination of the nucleic acid molecules described herein, including complements, analogs, variants, and fragments thereof, or polypeptides encoded by these nucleic acid molecules.
- the composition may include 2, 3, 4, 5, 6, 7, 8, 9, 10, etc. up to all of the nucleic acid molecules or polypeptides described herein, e.g., in any one or more of the Tables or Figures herein.
- the composition may include subsets of the nucleic acid molecules or polypeptides described herein, e.g., in any one or more of the Tables or Figures herein, for example, subsets groups by protein function or characteristics, e.g., proteins involved in the ubiquitination pathway, or proteins localized to a particular cellular compartment.
- nucleic acid molecules or polypeptides may for example be used with a substrate (e.g, a solid substrate or a liquid substrate) in a variety of applications, including the diagnosis of HCC, or monitoring the progression of HCC.
- a substrate e.g, a solid substrate or a liquid substrate
- addressable collection is meant a combination of nucleic acid molecules or polypeptides capable of being detected by, for example, the use of hybridization techniques or antibody binding techniques or by any other means of detection known to those of ordinary skill in the art.
- nucleic acid or “nucleic acid molecule” encompass both RNA (plus and minus strands) and DNA, including cDNA, genomic DNA, and synthetic (e.g., chemically synthesized) DNA.
- the nucleic acid may be double-stranded or single-stranded. Where single-stranded, the nucleic acid may be the sense strand or the antisense strand.
- a nucleic acid molecule may be any chain of two or more covalently bonded nucleotides, including naturally occurring or non-naturally occurring nucleotides, or nucleotide analogs or derivatives.
- RNA is meant a sequence of two or more covalently bonded, naturally occurring or modified ribonucleotides.
- RNA is meant a sequence of two or more covalently bonded, naturally occurring or modified deoxyribonucleotides.
- cDNA is meant complementary or copy DNA produced from an RNA template by the action of RNA-dependent DNA polymerase (reverse transcriptase).
- a cDNA clone means a duplex DNA sequence complementary to an RNA molecule of interest, carried in a cloning vector.
- An "oligonucleotide” as used herein is a single stranded molecule which may be used in hybridization or amplification technologies.
- an oligonucleotide may be any integer from about 15 to about 100 nucleotides in length, but may also be of greater length.
- a "probe” or “primer” is a single-stranded DNA or RNA molecule of defined sequence that can base pair to a second DNA or RNA molecule that contains a complementary sequence (the target). The stability of the resulting hybrid molecule depends upon the extent of the base pairing that occurs, and is affected by parameters such as the degree of complementarity between the probe and target molecule, and the degree of stringency of the hybridization conditions. The degree of hybridization stringency is affected by parameters such as the temperature, salt concentration, and concentration of organic molecules, such as formamide, and is determined by methods that are known to those skilled in the art.
- Probes or primers specific for the nucleic acid sequences described herein, or portions thereof may vary in length by any integer from at least 8 nucleotides to over 500 nucleotides, including any value in between, depending on the purpose for which, and conditions under which, the probe or primer is used.
- a probe or primer may be 8, 10, 15, 20, or 25 nucleotides in length, or may be at least 30, 40, 50, or 60 nucleotides in length, or may be over 100, 200, 500, or 1000 nucleotides in length.
- Probes or primers specific for the nucleic acid molecules described herein may have greater than any integer between 20-30% sequence identity, or at least any integer between 55- 75% sequence identity, or at least any integer between 75-85% sequence identity, or at least any integer between 85-99% sequence identity, or 100% sequence identity to the nucleic acid sequences described herein. Probes or primers can be detectably- labeled, either radioactively or non-radioactively, by methods that are known to those skilled in the art.
- Probes or primers can be used for methods involving nucleic acid hybridization, such as nucleic acid sequencing, nucleic acid amplification by the polymerase chain reaction, single stranded conformational polymorphism (SSCP) analysis, restriction fragment polymorphism (RFLP) analysis, Southern hybridization, northern hybridization, in situ hybridization, electrophoretic mobility shift assay (EMS A), microarray, and other methods that are known to those skilled in the art. Probes or primers may be derived from genomic DNA or cDNA, for example, by amplification, or from cloned DNA segments, or may be chemically synthesized.
- the "expression product" of a nucleic acid molecule may be any polypeptide encoded by that nucleic acid molecule. Generally, the polypeptide is capable of being expressed.
- a “protein,” “peptide” or “polypeptide” is any chain of two or more amino acids, including naturally occurring or non-naturally occurring amino acids or amino acid analogues, regardless of post-translational modification (e.g., glycosylation or phosphorylation).
- An "amino acid sequence”, “polypeptide”, “peptide” or “protein” of the invention may include peptides or proteins that have abnormal linkages, cross links and end caps, non-peptidyl bonds or alternative modifying groups. Such modified peptides are also within the scope of the invention.
- modifying group is intended to include structures that are directly attached to the peptidic structure (e.g., by covalent coupling), as well as those that are indirectly attached to the peptidic structure (e.g., by a stable non-covalent association or by covalent coupling to additional amino acid residues, or mimetics, analogues or derivatives thereof, which may flank the core peptidic structure).
- the modifying group can be coupled to the amino-terminus or carboxy-terminus of a peptidic structure, or to a peptidic or peptidomimetic region flanking the core domain.
- the modifying group can be coupled to a side chain of at least one amino acid residue of a peptidic structure, or to a peptidic or peptido- mimetic region flanking the core domain (e.g., through the epsilon amino group of a lysyl residue(s), through the carboxyl group of an aspartic acid residue(s) or a glutamic acid residue(s), through a hydroxy group of a tyrosyl residue(s), a serine residue(s) or a threonine residue(s) or other suitable reactive group on an amino acid side chain).
- a side chain of at least one amino acid residue of a peptidic structure or to a peptidic or peptido- mimetic region flanking the core domain (e.g., through the epsilon amino group of a lysyl residue(s), through the carboxyl group of an aspartic acid residue(s) or a glutamic acid residue(s), through a
- Modifying groups covalently coupled to the peptidic structure can be attached by means and using methods well known in the art for linking chemical structures, including, for example, amide, alkylamino, carbamate or urea bonds.
- Peptides according to the invention may include peptides encoded by the nucleic acid molecules of Tables 1 through 4 or complements or analogs thereof.
- differential expression or “differentially expressed” is meant increased, upregulated or present, or decreased, downregulated or absent, gene expression as detected by the absence, presence, or change (up or down) in the amount of transcribed messenger RNA or translated protein in a sample.
- the change may be detected by comparison of the change in gene expression level between a HCC sample and a non-tumor sample.
- the absolute amount of change of gene expression is not important, as long as the amount of change is reproducible, and measurable.
- the change (up or down) in the amount of transcribed messenger or translated protein may be at least 1-fold or at least 1.5-fold or maybe over 2.0, 2.5., 3.0, 3.5, 4.0, 4.5, or 5.0-fold.
- the change in the amount of transcribed messenger or translated protein may be 40%, 50%, 60%, 70%, 80%, 90%, or 100%.
- detecting it is intended to include determining the presence or absence, or quantifying the amount, of a nucleic acid molecule or polypeptide of the invention a substance.
- the term thus refers to the use of the materials, compositions, and methods of the present invention for qualitative and quantitative determinations.
- detecting an increase in gene expression levels may include quantifying a change of any value between 10% and 90%, or of any value between 30% and 60%, or over 100%, of any of the nucleic acid molecules or polypeptides of the invention when compared to a control.
- detecting an increase in gene expression levels may include quantifying a change of any value between 1 to 5 fold or more of any of the nucleic acid molecules or polypeptides of the invention when compared to a control.
- Hepatocellular carcinoma is cancer that arises from hepatocytes, the major cell type of the liver. It is a form of adenocarcinoma, and is the most common type of liver tumor.
- Non-tumor tissue refers to tissue or cells that are non-cancerous. In some embodiments, non-tumor tissue may include tissue or cells from a subject having a liver disorder, such as HBV or HCV infection, cirrhosis, exposure to aflatoxins, etc.
- the phrase "suspected of being cancerous" as used herein means a HCC tissue sample believed by one of ordinary skill in the art to contain HCC cells.
- non-cancerous or “non-tumor” is meant a tissue sample demonstrated by standard diagnostic or other techniques (e.g., histologic staining, microscopic analysis, immunoassay, etc.) to contain no HCC cells or evidence of HCC.
- a “sample” can be any organ, tissue, cell, or cell extract isolated from a subject, such as a sample isolated from a mammal having a hepatocellular carcinoma or isolated from a mammal not having a hepatocellular carcinoma or a tumor.
- a sample can include, without limitation, tissue such as liver tissue (e.g., from a biopsy or autopsy), cells, peripheral blood, whole blood, red cell concentrates, platelet concentrates, leukocyte concentrates, blood cell proteins, blood plasma, platelet-rich plasma, a plasma concentrate, a precipitate from any fractionation of the plasma, a supernatant from any fractionation of the plasma, blood plasma protein fractions, purified or partially purified blood proteins or other components, serum, semen, mammalian colostrum, milk, urine, stool, saliva, placental extracts, amniotic fluid, a cryoprecipitate, a cryosupernatant, a cell lysate, mammalian cell culture or culture medium, products of fermentation, ascitic fluid, proteins present in blood cells, solid tumours isolated from a mammal with a hepatocellular carcinoma, or any other specimen, or any extract thereof, obtained from a patient (human or animal), test subject, or experimental animal.
- tissue such as liver tissue (
- a sample may also include, without limitation, products produced in cell culture by normal, non-tumor, or transformed cells (e.g., via recombinant DNA technology).
- a "sample” may also be a cell or cell line created under experimental conditions, that are not directly isolated from a subject.
- a sample can also be cell-free, artificially derived or synthesised.
- samples refer to liver tissue or cells.
- the liver tissue may be from a subject having a hepatocellular carcinoma; a subject infected with a hepatitis virus; a subject having a liver disorder e.g., cirrhosis, or a subject having a normal liver e.g., not diagnosed with or suspected of having a liver disorder.
- a subject may be a human, non-human primate, rat, mouse, cow, horse, pig, sheep, goat, dog, cat, etc.
- the subject may be a clinical patient, a clinical trial volunteer, an experimental animal, etc.
- the subject may be suspected of having HCC, be diagnosed with HCC, or be a control subject that is confirmed to not have HCC.
- Diagnostic methods for HCC and the clinical delineation of HCC diagnoses are known to those of ordinary skill in the art, and include biopsy including radiological biopsy by means of a radiological scan, laproscopy, or open surgical biopsy.
- Figure 1 is a plot showing natural patterns of gene expression differences between HCC tumor and non-tumor liver tissue specimens based on unsupervised clustering. The plot shows the variance of expression value for each of the gene features across all the HCC tumor and non-tumor liver tissue specimens. The dotted line indicates the 500 most variable gene features.
- Figure 2 is a multidimensional scaling plot showing significant gene differential expression between HCC tumor and non-tumor liver tissues, and comparison with liver cancer cell lines (P ⁇ lxl0 " , approximately 1.5-fold change).
- the plot illustrates the ability of the 218 outlier genes to separate HCC tumor specimens (black circles) from non-tumor liver tissue specimens (dark gray circles).
- the plot also shows how different liver cancer cell lines (light gray circles) are from the clinical tissue samples.
- Figures 3A-B characterize differentially expressed genes in HCC tumor specimens (P ⁇ lxl0 "6 , approximately 1.5-fold change).
- Figure 3A is a bar graph showing the chromosomal distribution of the 218 outlier genes. The dark colored and light shaded bars represent genes that are at least 1.5-fold up- and downregulated, respectively, in HCC tumors relative to non-tumor livers.
- Figure 3B is a bar graph showing the functional characterization of the outlier genes based on Gene Ontology and published works.
- Figure 4 is a bar graph showing the expression of BMI-1 in HCC tumors as determined by cDNA microarray analysis. The data are presented as the level of expression (log base 2) in each HCC tumor specimen with respect to the corresponding non-tumor liver sample.
- Figures 5A-D show real-time RT-PCR analysis of IGFBP3, ERBB3, ERBB2 and EGFR in HCC tumor samples.
- the gene expression patterns for (A) IGFBP3, (B) ERBB3, (C) ERBB2 and (D) EGFR in all the 37 HCC tumor samples and their corresponding non-tumor liver tissue specimens were examined. All data was normalized to the amount of 'housekeeping' gene PBGD and are presented as relative fold expression change (log base 2 ratio) in HCC tumor specimens with respect to their corresponding non-tumor liver counterpart. Positive value depicts higher expression level, while negative value depicts lower expression level in the tumor relative to the non-tumor specimen.
- Figure 6 lists a panel of genes analyzed using real-time and semi-quantitative RT-PCR analyses, and indicating whether the analysis was conducted in non-rumor human tissues, and in clinical tissue samples or HCC cell lines or both.
- Figures 7A-C show gene expression analysis of ARMET.
- Semi-quantitative RT-PCR analysis (A) of non-tumor tissues and HCC cell lines and real time RT-PCR analysis of non-tumor tissues (B) and patient samples (C) was performed.
- GAPDH expression level was used as the control in the analyses of HCC cell lines vs. non- tumor human tissues (A).
- the dotted line in (B) indicates mean expression value of four non-tumor liver tissues i.e., Fetal/F, Fetal/M, Adult/F, Adult/M.
- Figures 8 A-C show gene expression analysis of BMI-1.
- RT-PCR analysis A of non-tumor tissues and HCC cell lines and real time RT-PCR analysis of non-tumor tissues (B) and patient samples (C) was performed.
- GAPDH expression level was used as the control in the analyses of HCC cell lines vs. non- tumor human tissues (A).
- the dotted line in (B) indicates mean expression value of four non-tumor liver tissues i.e., Fetal/F, Fetal/M, Adult/F, Adult/M.
- Figures 9A-C show gene expression analysis of CRHBP.
- Semi-quantitative RT-PCR analysis (A) of non-tumor tissues and HCC cell lines and real time RT-PCR analysis of non-tumor tissues (B) and patient samples (C) was performed.
- GAPDH expression level was used as the control in the analyses of HCC cell lines vs. non- tumor human tissues (A).
- the dotted line in (B) indicates mean expression value of four non-tumor liver tissues i.e., Fetal/F, Fetal/M, Adult/F, Adult/M.
- Figures 10A-C show gene expression analysis of CSTB.
- Semi-quantitative RT-PCR analysis (A) of non-tumor tissues and HCC cell lines and real time RT-PCR analysis of non-tumor tissues (B) and patient samples (C) was performed.
- GAPDH expression level was used as the control in the analyses of HCC cell lines vs. non- tumor human tissues (A).
- the dotted line in (B) indicates mean expression value of four non-tumor liver tissues i.e., Fetal F, Fetal/M, Adult F, Adult M.
- Figures 11 A-C show gene expression analysis of DPT.
- Semi-quantitative RT-PCR analysis (A) of non-tumor tissues and HCC cell lines and real time RT-PCR analysis of non-tumor tissues (B) and patient samples (C) was performed.
- GAPDH expression level was used as the control in the analyses of HCC cell lines vs. non- tumor human tissues (A).
- the dotted line in (B) indicates mean expression value of four non-tumor liver tissues i.e., Fetal/F, Fetal/M, Adult/F, Adult/M.
- Figures 12A-B show gene expression analysis of ERBB3.
- Real time RT-PCR analysis of non-tumor tissues (A) and patient samples (B) was performed.
- the dotted line in (A) indicates mean expression value of four non-tumor liver tissues i.e., Fetal/F, Fetal/M, Adult F, Adult/M.
- Figures 13A-B show gene expression analysis of EZH2. Real time RT-PCR analysis of non-tumor tissues (A) and patient samples (B) was performed.
- the dotted line in (A) indicates mean expression value of four non-tumor liver tissues i.e., Fetal/F, Fetal/M, Adult/F, Adult/M.
- Figures 14A-B show gene expression analysis of GPC3.
- Real time RT-PCR analysis of non-tumor tissues (A) and patient samples (B) was performed.
- the dotted line in (A) indicates mean expression value of four non-tumor liver tissues i.e., Fetal/F, Fetal/M, Adult/F, Adult/M.
- Figures 15A-B show gene expression analysis of HDGF.
- Real time RT-PCR analysis of non-tumor tissues (A) and patient samples (B) was performed.
- the dotted line in (A) indicates mean expression value of four non-tumor liver tissues i.e., Fetal/F, Fetal/M, Adult/F, Adult/M.
- Figures 16A-B show gene expression analysis of MDK. Real time RT-PCR analysis of non-tumor tissues (A) and patient samples (B) was performed. The dotted line in (A) indicates mean expression value of four non-tumor liver tissues i.e., Fetal/F, Fetal/M, Adult/F, Adult M.
- Figure 17 shows gene expression analysis of D123. Semi-quantitative RT- PCR analysis of non-tumor tissues and HCC cell lines was performed. GAPDH expression level was used as the control in the analyses of HCC cell lines vs. non- tumor human tissues.
- Figure 18 shows gene expression analysis of FLJ10326. Semi-quantitative RT-PCR analysis of non-tumor tissues and HCC cell lines was performed. GAPDH expression level was used as the control in the analyses of HCC cell lines vs. non- tumor human tissues.
- Figure 19 shows gene expression analysis of ICA-1A. Semi-quantitative RT- PCR analysis of non-tumor tissues and HCC cell lines was performed. GAPDH expression level was used as the control in the analyses of HCC cell lines vs. non- tumor human tissues.
- Figure 20 shows gene expression analysis of LASP1.
- Semi-quantitative RT- PCR analysis of non-tumor tissues and HCC cell lines was performed.
- GAPDH expression level was used as the control in the analyses of HCC cell lines vs. non- tumor human tissues.
- Figure 21 shows gene expression analysis of PODXL. Semi-quantitative RT- PCR analysis of non-tumor tissues and HCC cell lines was performed. GAPDH expression level was used as the control in the analyses of HCC cell lines vs. non- tumor human tissues.
- Phenotypic changes in cancer may be due to cellular changes at the nucleotide level. Thus, some genes may be expressed, overexpressed, or under-expressed in tumor cells relative to non-tumor cells.
- Selecting a set of differentially expressed HCC genes, nucleic acid molecules, and/or polypeptides assists in predictable and accurate diagnosis and therapy, and design of efficacious therapeutics.
- the invention provides, in part, nucleic acid molecules and polypeptides that are differentially expressed in HCC cells, when compared to non-HCC tissue, e.g., liver or serum.
- non-HCC tissue e.g., liver or serum.
- the invention provides, in part, molecular markers for HCC derived from the analysis of global changes in gene expression ("gene expression profiles") between HCC tissue and non-HCC tissue. More specifically, cDNA microarrays were used to examine the global cellular changes in matched pairs of HCC tumor and non-tumor tissues of patients diagnosed with HCC. In addition, gene expression patterns between primary HCC tumors and liver cancer cell lines were examined for possible biological variation.
- the nucleic acid molecules or polypeptides provided by the invention serve as molecular markers that may be used for example for HCC diagnosis; to assess HCC progression or regression; to access the efficacy and/or toxicity of HCC therapeutics; and/or to identify candidate compounds for HCC therapy, with high predictive accuracy.
- the genes lists identified permit rapid, simple, and reproducible screening of a variety of HCC samples by, for example, nucleic acid microarray hybridization or protein expression technology to determine the expression of the specific genes, or by other means such as differential display, gel electrophoresis, genome mismatch scanning, representational discriminate analysis, clustering, transcript imaging, etc. used singly or in combination.
- the selected nucleic acid molecules or polypeptides of the invention define standard and reproducible differential expression patterns against which to compare the expression pattern in a variety of tissue or cells, e.g., HCC tissue or cells or serum, obtained by biopsy, autopsy, or from cell lines and/or in vitro treatment or assays.
- the selected nucleic acid molecules or polypeptides of the invention and subsets thereof provide reliable detection of HCC cells or tissue, with reduction or elimination of false positives or false negatives.
- the invention provides composite sets of discriminator genes for use as general or global HCC tumor markers.
- the nucleic acid molecules or polypeptides of the invention may be used to assess the suitability of a HCC cell line for use as a model for HCC, as gene expression profiles may vary between primary HCC tumors and HCC cell lines.
- Compounds according to the invention include, without limitation, molecules substantially identical to the nucleic acid molecules of Tables 1 through 4 (e.g., BMI- 1, ARMET, CRHBP, CSTB, DPT, ERBB3, EZH2, GPC3, HDGF, MDK, D123, FLJ10326, ICAP-IA, LASP1, PODXL) and complements, analogs, fragments, and variants thereof, including, for example, the polypeptides described herein that are encoded by the nucleic acid molecules of Tables 1 through 4, as well as homologs and fragments thereof.
- compounds of the invention include antibodies that specifically bind to polypeptides encoded by the nucleic acid molecules of Tables 1 through 4.
- an antibody "specifically binds" an antigen when it recognises and binds the antigen, for example, a polypeptide encoded by any of the nucleic acid molecules described herein, but does not substantially recognise and bind other reference molecules in a sample, for example, a polypeptide that is encoded by a nucleic acid molecule that is not substantially identical to any of the nucleic acid molecules described herein.
- Such an antibody has, for example, an affinity for the antigen which is at least 10, 100, 1000 or 10000 times greater than the affinity of the antibody for another reference molecule in a sample.
- a "substantially identical" sequence is an amino acid or nucleotide sequence that differs from a reference sequence only by one or more conservative substitutions, as discussed herein, or by one or more non-conservative substitutions, deletions, or insertions located at positions of the sequence that do not destroy the biological function of the amino acid or nucleic acid molecule, or that do not destroy the detectability (e.g., by hybridization or specific binding) of the amino acid or nucleic acid molecule.
- Such a sequence can be any integer from 10% to 99%, or more generally at least 10%, 20%, 30%, 40%, 50, 55% or 60%, or at least 65%, 75%, 80%, 85%, 90%, or 95%, or as much as 96%, 97%, 98%, or 99% identical when optimally aligned at the amino acid or nucleotide level to the sequence used for comparison using, for example, the Align Program (Myers and Miller, CABIOS, 1989, 4:11-17) or FAST A.
- the length of comparison sequences may be at least 2, 5, 10, or 15 amino acids, or at least 20, 25, or 30 amino acids.
- the length of comparison sequences maybe at least 35, 40, or 50 amino acids, or over 60, 80, or 100 amino acids.
- the length of comparison sequences maybe at least 5, 10, 15, 20, or 25 nucleotides, or at least 30, 40, or 50 nucleotides.
- the length of comparison sequences maybe at least 60, 70, 80, or 90 nucleotides, or over 100, 200, or 500 nucleotides.
- Sequence identity can be readily measured using publicly available sequence analysis software (e.g., Sequence Analysis Software Package of the Genetics Computer Group, University of Wisconsin Biotechnology Center, 1710 University Avenue, Madison, Wis. 53705, or BLAST software available from the National Library of Medicine, or as described herein).
- Examples of useful software include the programs Pile-up and PrettyBox. Such software matches similar sequences by assigning degrees of homology to various substitutions, deletions, substitutions, and other modifications. Alternatively, or additionally, two nucleic acid sequences may be "substantially identical" if they hybridize under high stringency conditions.
- high stringency conditions are, for example, conditions that allow hybridization comparable with the hybridization that occurs using a DNA probe of at least 500 nucleotides in length, in a buffer containing 0.5 M NaHPO 4 , pH 7.2, 7% SDS, 1 mM EDTA, and 1% BSA (fraction V), at a temperature of 65°C, or a buffer containing 48% formamide, 4.8x SSC, 0.2 M Tris-Cl, pH 7.6, lx Denhardt's solution, 10% dextran sulfate, and 0.1% SDS, at a temperature of 42°C.
- Hybridizations maybe carried out over a period of about 20 to 30 minutes, or about 2 to 6 hours, or about 10 to 15 hours, or over 24 hours or more.
- High stringency hybridization is also relied upon for the success of numerous techniques routinely performed by molecular biologists, such as high stringency PCR, DNA sequencing, single strand conformational polymorphism analysis, and in situ hybridization. In contrast to northern and Southern hybridizations, these techniques are usually performed with relatively short probes (e.g., usually about 16 nucleotides or longer for PCR or sequencing and about 40 nucleotides or longer for in situ hybridization).
- a "variant" is a nucleic acid molecule that is a recognized variation of a nucleic acid molecule or expression product thereof. Splice variants may be determined for example by using computer programs, e.g, BLAST. Allelic variants have in general a high percent identity to the nucleic acid molecule of interest.
- Single nucleotide polymorphism refers to a change in a single base as a result of a substitution, insertion or deletion. The change may be conservative (purine for purine) or non-conservative (purine to pyrimidine) and may or may not result in a change in an encoded amino acid.
- An "analog” is a nucleic acid molecule or polypeptide that has been subjected to a chemical modification. Nucleic acid analogs can include substitution of a non-traditional base such as queosine or of an analog such as hypoxanthine, or other substitutions known in the art. Analogs in general retain the biological activities of the naturally occurring molecules but may confer advantages such as longer lifespan or enhanced activity.
- nucleic acids e.g., DNA or RNA
- nucleic acids e.g., DNA or RNA
- each nucleotide in a nucleic acid molecule need not form a matched Watson- Crick base pair with a nucleotide in an opposing complementary strand to form a duplex.
- a nucleic acid molecule is "complementary" to another nucleic acid molecule, or is a “complement” of that other nucleic acid molecule, if it hybridizes, under conditions of high stringency, with the second nucleic acid molecule.
- the "complement” of a nucleic acid molecule of Tables 1 through 4 may in some embodiments include a nucleic acid molecule that is complementary over the full length of the sequence of a nucleic acid molecule of Tables 1 through 4.
- a “fragment” may be any portion of a nucleic acid molecule or polypeptide as described herein that is capable of being differentially expressed or detected in an assay or screening method according to the invention.
- genes and nucleic acid sequences of the invention maybe recombinant sequences.
- the term "recombinant” means that something has been recombined, so that when made in reference to a nucleic acid construct the term refers to a molecule that is comprised of nucleic acid sequences that are joined together or produced by means of molecular biological techniques.
- the term "recombinant” when made in reference to a protein or a polypeptide refers to a protein or polypeptide molecule which is expressed using a recombinant nucleic acid construct created by means of molecular biological techniques.
- Recombinant nucleic acid constructs may include a nucleotide sequence which is ligated to, or is manipulated to become ligated to, a nucleic acid sequence to which it is not ligated in nature, or to which it is ligated at a different location in nature. Referring to a nucleic acid construct as 'recombinant' therefore indicates that the nucleic acid molecule has been manipulated using genetic engineering, i.e. by human intervention. Recombinant nucleic acid constructs may for example be introduced into a host cell by transformation.
- Such recombinant nucleic acid constructs may include sequences derived from the same host cell species or from different host cell species, which have been isolated and reintroduced into cells of the host species. Recombinant nucleic acid construct sequences may become integrated into a host cell genome, either as a result of the original transformation of the host cells, or as the result of subsequent recombination and/or repair events.
- heterologous in reference to a nucleic acid or protein is a molecule that has been manipulated by human intervention so that it is located in a place other than the place in which it is naturally found.
- a nucleic acid sequence from one species may be introduced into the genome of another species, or a nucleic acid sequence from one genomic locus may be moved to another genomic or extrachromasomal locus in the same species.
- a heterologous protein includes, for example, a protein expressed from a heterologous coding sequence or a protein expressed from a recombinant gene in a cell that would not naturally express the protein.
- a compound is "substantially pure" when it is separated from the components that naturally accompany it.
- a compound is substantially pure when it is at least 10%, 20%, 30%, 40%, 50%, or 60%, more generally 70%, 75%, 80%, or 85%, or over 90%, 95%, or 99% by weight, of the total material in a sample.
- a polypeptide that is chemically synthesised, produced by recombinant technology, isolated by known purification techniques will be generally be substantially free from its naturally associated components.
- a substantially pure compound can be obtained, for example, by extraction from a natural source; by expression of a recombinant nucleic acid molecule encoding a polypeptide compound; or by chemical synthesis.
- a nucleic acid molecule is substantially pure or "isolated" when it is not immediately contiguous with (i.e., covalently linked to) the coding sequences with which it is normally contiguous in the naturally occurring genome of the organism from which the DNA of the invention is derived.
- an "isolated" gene or nucleic acid molecule is intended to mean a gene or nucleic acid molecule which is not flanked by nucleic acid molecules which normally (in nature) flank the gene or nucleic acid molecule (such as in genomic sequences) and/or has been completely or partially purified from other transcribed sequences (as in a cDNA or RNA library).
- an isolated nucleic acid of the invention maybe substantially isolated with respect to the complex cellular milieu in which it naturally occurs.
- the isolated material will form part of a composition (for example, a crude extract containing other substances), buffer system or reagent mix.
- the material may be purified to essential homogeneity, for example as determined by PAGE or column chromatography such as HPLC.
- the term therefore includes, e.g., a recombinant nucleic acid incorporated into a vector, such as an autonomously replicating plasmid or virus; or into the genomic DNA of a prokaryote or eukaryote, or which exists as a separate molecule (e.g., a cDNA or a genomic DNA fragment produced by PCR or restriction endonuclease treatment) independent of other sequences. It also includes a recombinant nucleic acid which is part of a hybrid gene encoding additional polypeptide sequences.
- an isolated nucleic acid comprises at least about 40%, 50%, 60%, 70%, 80%, 90%, 95%, or 99% (on a molar basis) of all. macromolecular species present.
- an isolated gene or nucleic acid molecule can include a gene or nucleic acid molecule which is synthesized chemically or by recombinant means. Recombinant DNA contained in a vector are included in the definition of "isolated” as used herein.
- isolated nucleic acid molecules include recombinant DNA molecules in heterologous host cells, as well as partially or substantially purified DNA molecules in solution. In vivo and in vitro RNA transcripts of the DNA molecules of the present invention are also encompassed by "isolated" nucleic acid molecules.
- Such isolated nucleic acid molecules are useful in the manufacture of the encoded polypeptide, as probes for isolating homologous sequences (e.g., from other mammalian species), for gene mapping (e.g., by in situ hybridization with chromosomes), or for detecting expression of the gene in tissue (e.g., human tissue, such as peripheral blood), such as by Northern blot analysis.
- Polypeptide compounds can be prepared by, for example, replacing, deleting, or inserting an amino acid residue at any position of a peptide or a peptide analog, for example, a peptide as described herein, with other conservative amino acid residues, i.e., residues having similar physical, biological, or chemical properties.
- polypeptides of the present invention also extend to biologically equivalent peptides that differ from a portion of the sequence of the polypeptides of the present invention by conservative amino acid substitutions.
- conservative amino acid substitutions refers to the substitution of one amino acid for another at a given location in the peptide, where the substitution can be made without substantial loss of the relevant function.
- substitutions of like amino acid residues can be made on the basis of relative similarity of side-chain substituents, for example, their size, charge, hydrophobicity, hydrophilicity, and the like, and such substitutions may be assayed for their effect on the function of the peptide by routine testing.
- Conservative changes can also include the substitution of a chemically derivatised moiety for a non-derivatised residue, by for example, reaction of a functional side group of an amino acid.
- Peptides or peptide analogs can be synthesised by standard chemical techniques, for example, by automated synthesis using solution or solid phase synthesis methodology. Automated peptide synthesisers are commercially available and use techniques well known in the art.
- Peptides and peptide analogs can also be prepared using recombinant DNA technology using standard methods such as those described in, for example, Sambrook, et al. (Molecular Cloning: A Laboratory Manual. 2 nd ed., Cold Spring Harbor Laboratory, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y., 1989) or Ausubel et al. (Current Protocols in Molecular Biology, John Wiley & Sons, 1994).
- candidate HCC therapeutics may also be advantageous when screening candidate HCC therapeutics.
- candidate compounds are screened and prescreened for the ability to interact with a major target without regard to other effects they may have on cells or in the subject to be treated, such as toxicity, which prevent the development and use of the potential compound.
- the methods of the invention may be used to identify candidate compounds suitable for HCC therapy.
- candidate or test compounds are identified from large libraries of both natural products or synthetic (or semi-synthetic) extracts or chemical libraries according to methods known in the art.
- test extracts or compounds is not critical to the methods of the invention. Accordingly, virtually any number of chemical extracts or compounds can be screened using the exemplary methods described herein.
- extracts or compounds include, but are not limited to, plant-, fungal-, prokaryotic- or animal-based extracts, fermentation broths, and synthetic compounds, as well as modification of existing compounds. Numerous methods are also available for generating random or directed synthesis (e.g., semi- synthesis or total synthesis) of any number of chemical compounds, including, but not limited to, saccharide-, lipid-, peptide-, and nucleic acid-based compounds. Synthetic compound libraries are commercially available. Alternatively, libraries of natural compounds in the form of bacterial, fungal, plant, and animal extracts are commercially available from a number of sources, including Biotics (Sussex, UK), Xenova (Slough, UK), Harbor Branch Oceanographic Institute (Ft.
- Candidate compounds useful for treating HCC may also be identified by assessing variations in the expression of one or more HCC markers, from Tables 1 through 4, prior to and after contacting HCC cells or tissues with candidate pharmacological agents for the treatment of HCC.
- the cells may be grown in culture (e.g. from a HCC cell line), or may be obtained from a subject, (e.g. in a clinical trial of candidate pharmaceutical agents to treat HCC).
- HCC nucleic acid markers drug targets
- a candidate pharmacological agent to treat HCC indicates progression, regression, or stasis of the HCC thereby indicating efficacy of candidate agents and concomitant identification of candidate compounds for therapeutic use in HCC.
- Candidate compounds may also be screened for toxicity, specificity, etc.
- the invention provides nucleic acid or polypeptide arrays and biological assays thereof.
- Arrays refer to ordered arrangements of at least two nucleic acid molecules or polypeptides on a substrate, which can be any rigid or semi-rigid support to which two nucleic acid molecules or polypeptides may be attached.
- a substrate may be a liquid medium.
- Substrates include membranes, filters, chips, slides, wafers, fibers, beads, gels, capillaries, plates, polymers, and microparticles etc.
- the hybridization or binding patterns and intensities create a unique expression profile, which can be interpreted in terms of expression levels of particular genes and can be correlated with HCC progression, regression, therapy, etc.
- High density nucleic acid or polypeptide arrays are also referred to as "microarrays," and may for example be used to monitor the presence or level of expression of a large number of genes or polypeptides or for detecting sequence variations, mutations and polymorphisms.
- Arrays and microarrays generally require a solid support (for example, nylon, glass, ceramic, plastic, silica, aluminosilicates, borosilicates, metal oxides such as alumia and nickel oxide, various clays, nitrocellulose, etc.) to which the nucleic acid molecules or polypeptides are attached in a specified 2-dimensional arrangement, such that the pattern of hybridization or binding to a probe is easily determinable.
- At least one of the nucleic acid molecules or polypeptides is a control, standard, or reference molecule, such as a housekeeping gene or portion thereof (e.g., PBGD, GAPDH), that may assist in the normalization of expression levels or assist in the determining of nucleic acid quality and binding characteristics; reagent quality and effectiveness; hybridization success; analysis thresholds and success, etc.
- a housekeeping gene or portion thereof e.g., PBGD, GAPDH
- Nucleic acid molecules or polypeptide probes may be derived from compounds as described herein for example in Tables 1 through 4, and the compositions of the invention may be used as elements on a microarray to analyze gene expression profiles.
- nucleic acids may include any polymer or oligomer of nucleosides or nucleotides (polynucleotides or oligonucleotides), which include pyrimidine and purine bases, preferably cytosine, thymine, and uracil, and adenine and guanine, respectively.
- pyrimidine and purine bases preferably cytosine, thymine, and uracil, and adenine and guanine, respectively.
- a variety of methods are known for making and using microarrays, as for example disclosed in Cheung, V.G., et al.
- the microarray substrate may be coated with a compound to enhance synthesis of the nucleic acid molecule on the substrate as disclosed in, for example, U.S. Pat. No.4,458,066.
- probes may be synthesized directly on the substrate in a predetermined ordered arrangement.
- the invention provides nucleic acid or polypeptide microarrays including a number of distinct and selected nucleic acid or polypeptide array sequences of the invention.
- the number of distinct sequences may for example be any integer between 2 and 1 x 10 5 , such as at least 10 2 , 10 3 , 10 4 , or 10 5 .
- the size of the distinct sequences may vary depending on the intended use, and can be determined by a skilled person.
- the nucleic acid sequences may range from 15 to 5000 bases or more, or any integer between this range.
- Microarrays may also be used to examine the expression of all the genes in a tissue or cell such as a liver cell or a HCC cell.
- the nucleic acid molecules of Tables 1 through 4 may be attached to a solid support, hybridized with single stranded detectably-labeled cDNAs (corresponding to a "complementary" orientation), and quantified using an appropriate method such that a signal is detected at each location at which hybridization has taken place. The intensity of the signal would then reflect the amount of gene expression.
- protein microarrays may be used according to methods known in the art.
- hepatocellular carcinoma cells or tissue for example, hepatocellular carcinoma cells or tissue, hepatitis virus infected cells or tissue, non-tumor cells or tissue, normal cells or tissue, cirrhotic liver cells or tissue, or any combination thereof would elucidate differing levels of expression of specified genes from the different sources.
- libraries maybe constructed of bacterial strains each of which bears a plasmid expressing a different nucleic acid molecule of any one or more of Tables 1 through 4 under control of an inducible promoter.
- ORFs are amplified using PCR and cloned into a vector that enables their expression as N- terminal his-tagged polypeptides. These amplicons are also used to construct hybridization microarrays and enable targeted gene disruption, reducing expenses.
- a suitable expression host e.g. E. coli
- genes encoding particular biochemical activities are identified by screening arrayed pools of his-tagged proteins as described previously (Martzen, M.R., et al., 1999).
- the invention also provides databases including the nucleic acid and polypeptide sequences described herein, as well as gene expression information in various cancerous and non-cancerous liver and liver cell line samples. Such databases may be used to access information that may aid in diagnosis, prognosis, or other HCC-related methods of the invention.
- a database as used herein includes any electronic form of the compounds (e.g., nucleic acid and polypeptide sequences) of the invention, and information regarding these compounds, and includes computer readable media and any suitable form for storing the information.
- kits including for example one or more of the nucleic acid molecules or polypeptides of the invention (or complements, analogs, variants, or fragments thereof), an appropriate buffer, appropriate reagents for detection, and appropriate controls.
- a kit may include probes or primers (which may or may not be detectably labeled) suitable for hybridization or amplification, or may include antibodies or ligands suitable for specific binding.
- a kit may also include written or electronic instructions.
- detectable labels and conjugation techniques are known by those skilled in the art and may be used in various nucleic acid molecule and polypeptide assays to diagnose HCC.
- the nucleic acid molecules, proteins, antibodies and other compounds according to the invention may be labeled for purposes of assay by joining them, either covalently or noncovalently, with a detectable label.
- detectably labeled is meant any means for marking and identifying the presence of a molecule, e.g., an.oligonucleotide probe or primer, a gene or fragment thereof, a cDNA molecule, or a polypeptide.
- Methods for detectably-labeling a molecule include, without limitation, radioactive labeling (e.g., with an isotope such as 32 P or 35 S) and nonradioactive labelling such as, enzymatic labeling (for example, using horseradish peroxidase or alkaline phosphatase), chemiluminescent labeling, fluorescent labeling (for example, using fluorescein), bioluminescent labeling, or antibody detection of a ligand attached to the probe.
- radioactive labeling e.g., with an isotope such as 32 P or 35 S
- nonradioactive labelling such as, enzymatic labeling (for example, using horseradish peroxidase or alkaline phosphatase), chemiluminescent labeling, fluorescent labeling (for example, using fluorescein), bioluminescent labeling, or antibody detection of a ligand attached to the probe.
- a molecule that is detectably labeled by an indirect means for example, a molecule that is bound with a first moiety (such as biotin) that is, in turn, bound to a second moiety that may be observed or assayed (such as fluorescein-labeled streptavidin).
- Labels also include digoxigenin, luciferases, and aequorin. Synthesis of labeled molecules performed by using labels such as 32 P-dCTP, Cy3-dCTP or Cy5-dCTP or 35 S-methionine.
- Compounds according to the invention may also be directly labeled by chemical conjugation to amines, thiols and other groups present in the molecules using reagents such as BIODEPY or FITC (Molecular Probes, Eugene, OR, USA).
- Compounds, compositions, and reagents according to the invention may be used to detect and quantify differential gene expression; absence, presence, or excess expression of nucleic acid molecules (e.g., mRNAs) or polypeptides; or to monitor nucleic acid molecule (e.g., mRNA) or polypeptide levels during therapeutic intervention in subjects with HCC.
- the compounds, compositions, and reagents according to the invention can also be utilized as markers of HCC treatment efficacy over a period ranging from days to months to years.
- the diagnostic assays may use hybridization, amplification, ligand binding, or antibody technologies to compare gene expression in a biological sample from a subject to reference samples or standards, or to cancerous and non-cancerous samples from the subject, in order to detect altered gene expression.
- Qualitative or quantitative methods for this comparison are known in the art, and any suitable method may be used.
- a non-tumor or standard gene expression profile may be established. This may be accomplished by combining a biological sample taken from normal or non-tumor subjects or from non-cancerous tissue from a subject with HCC, with a probe under conditions for hybridization or amplification.
- Standard hybridization may be quantified by comparing the values obtained using non-tumor subjects or non-cancerous tissue with values from an experiment in which a known amount of a substantially purified target sequence is used. Standard values obtained in this manner may be compared with values obtained from samples from patients who are symptomatic for HCC. Deviation from standard values toward those associated with HCC is used to diagnose HCC. Such assays may also be used to monitor the efficacy of a particular HCC therapy in animal studies, in clinical trials, or to monitor the treatment of an individual patient or groups of patients.
- assays according to the invention may be repeated on a regular basis to determine if the level of expression in the subject begins to approximate that which is observed in a non-tumor subject, and to monitor the progression of HCC in the subject.
- the results obtained from successive assays may be used to show the efficacy of treatment over a period ranging from several days to months.
- Compounds, compositions, and reagents (e.g., microarrays) according to the invention may be used to monitor the progression or regression of HCC.
- the differences in gene expression between healthy and diseased tissues or cells can be assessed and cataloged.
- HCC By analyzing changes in patterns of gene expression, HCC can be diagnosed at earlier stages before the subject is symptomatic. Similarly, by analyzing gene expression profiles and changes therein, prognoses may be formulation, and therapies may be designed. Progression or regression of HCC may be determined by comparison of two or more different HCC samples taken at multiple different times from a subject (e.g., at least 2, 3, 4, or 5 or more time points) over the course of days to months. For example, progression or regression may be evaluated by assessments of expression of sets of two or more, or as many as all, of the nucleic acid molecules of Tables 1 through 4 in a HCC tissue sample from a subject before, during, and following treatment for HCC.
- Compounds, compositions, and reagents (e.g., microarrays) according to the invention can also be used to monitor the efficacy of a therapy.
- compounds, compositions, and reagents (e.g., microarrays) according to the invention may be employed to improve the therapeutic regimen.
- dosages that causes changes in gene profiles that represent efficacious treatment may be determined, and expression profiles associated with the onset of undesirable side effects may be avoided. This approach may be more sensitive and rapid than waiting for the subject to show inadequate improvement, or to manifest side effects, before altering the course of treatment.
- pre- and post-treatment alterations in expression of two or more sets of HCC nucleic acid molecules in HCC cells or tissues may be used to assess treatment parameters including, but not limited to: dosage, method of administration, timing of administration, and combination with other known treatments for HCC.
- any one or more of the compounds provided herein may be used in therapeutic applications.
- selected compounds provided herein maybe used as therapeutic targets for the identification of agents, that modulate their expression levels and/or activity, that may be used to treat HCC.
- EXAMPLES Experimental Procedures RNA isolation, RNA amplification and cDNA microarray hybridization
- RNA samples of tumor and corresponding non-tumor tissues were obtained from resected liver specimens from thirty-seven (37) patients who had been diagnosed with hepatitis B virus (HBV)-associated HCC and had undergone curative liver resection.
- a validation tissue set composed of 58 liver biopsy samples from an independent cohort of twenty-nine (29) patients, who also had HBV-associated HCC and had undergone liver resection, was used. Informed consent from the patient and institutional research and ethics committee approval were obtained. Tissues were snap frozen in liquid nitrogen and stored at -150°C. A small section of each specimen was sampled and total RNA was isolated from tissues using TRIZOL ® reagent (Life Technologies, Bethesda, MD, USA) according to the manufacturer's instructions. The integrity of the RNA specimen was verified by gel electrophoresis.
- the human liver cancer cell lines used in this study were: PLC/PRF/5,
- the cDNA was made blunt-ended with T4 DNA polymerase, and purified by extraction in a mixture of phenol, chloroform and isoamyl alcohol, and precipitation in the presence of ammonium acetate and ethanol. Purified double-stranded cDNA was then transcribed with T7 polymerase (T7 Megascript ® Kit, Ambion) to yield linearly amplified antisense RNA, which was subsequently purified with RNeasy ® rnini-columns (Qiagen). Human universal reference RNA (Stratagene, La Jolla, CA), including total RNA from 10 different human cell lines, was amplified and used as the reference for cDNA , microarray analysis.
- RNA samples were spotted onto poly-L-lysine coated slides using OmniGrid ® arrayer (GeneMachines). Probes were generated from the amplified RNA material and hybridized to the chip as described elsewhere (Sotiriou et al, 2002). Briefly, 4 ⁇ g amplified RNA was reverse-transcribed using random hexamers and directly labeled with Cy3 -conjugated dUTP (reference RNA) or Cy5-conjugated dUTP (sample RNA). Hybridization was performed in the presence of 25% formamide and 5X SSC for 16h at 42°C. Slides were scanned with an Axon 4000b laser scanner (Axon Instruments) after washing and drying. To minimize the effects of labeling biases, reciprocal dye swap labeling experiments were performed for each sample.
- the 37 paired HCC tumor and non-tumor liver samples, and liver cancer cell lines were processed on the microarray on two separate prints, and the validation tissue set was processed on a third print.
- Raw data was analyzed on GenePix analysis software version 3.0 (Axon Instruments, Burlingame, CA, USA) and uploaded to a relational database maintained by the Center for Information Technology at the National Institutes of Health (ie. MADB).
- the cDNA clones used for the microarray are represented by their UniGene identifiers. For each array, the logarithmic expression ratio for a spot on each array was normalized by subtracting the median logarithmic ratio for the same array.
- the quality of a set of selected gene features to be used as potential markers was measured by estimating the probability that its observed performance, in terms of number of misclassified tissue samples, could occur by chance alone. This was achieved by performing a series of Monte Carlo simulations (Davison and Hinkley, 1997) upon the expression data of the selected genes. In each simulation, the tissues' labels were randomly permuted and the number of misclassifications was noted. A total of one million runs of Monte Carlo simulations were performed. The reported P-value (denoted as P a ) is the fraction of times the permutations generated as few misclassifications as, or fewer than, the original labeling.
- each discriminator cassette was assessed on an independent tissue set comprising of 58 liver clinical biopsies from 29 patients using a ⁇ -Nearest Neighbor (£NN) classification algorithm using Pearson correlation to measure the similarity between expression profiles.
- £NN ⁇ -Nearest Neighbor
- RNA from individual tissue samples were analyzed for the expression levels of selected genes by real-time semi-quantitative RT-PCR using the LightCycler RNA amplification kit SYBR Green I on the LightCycler (Roche, Basel, Switzerland) according to the manufacturer's instructions. Briefly, one-step RT-PCR reactions consisted of an initial incubation at 55°C for 10 min, followed by a denaturation step at 95°C for 30 s, and amplification for 40 cycles of 1 s at 95°C, 10 s at 57°C, and 13 s at 72°C. For each reaction, 10 ng of total RNA was analyzed.
- the gene specific primers designed were, for example, as follows: IGFBP3 5'- ATAATCATCATCAAGAAAGGGCAT-3 ' and 5'-GAAGGGCGACACTGCTT- 3 '; EGFR 5'-GCGTCTCTTGCCGGAATG-3' and 5'-GGCTCACCCTCCAGAAGCTT- 3'; ERBB2 5'-GGATGTGCGGCTCGTACAC-3' and 5'- TAATTTTGACATGGTTGGGACTCTT- 3'; ERBB3 5'- CGGTTATGTCATGCCAGATACAC-3' and 5'- ACAGAACTGAGACCCACTGAAGAA-3 '; PBGD 5'-
- the gene expression patterns of primary HCC tumors and the corresponding non-tumor liver tissues from 37 patients were examined by cDNA microarray.
- Amplified RNA prepared from each experimental sample was labeled with Cy5 and hybridized on the array with pooled human 'common reference' amplified RNA labeled with Cy3. Reciprocal dye swap replicate hybridizations were performed to minimize technical noise. Since the overall correlation of reciprocal labeling was good, values obtained from reciprocal labeling experiments were averaged and used in subsequent analyses. Firstly, the overall natural patterns of gene expression in the HCC tumor and non-tumor liver tissues were assessed based on unsupervised hierarchical clustering.
- outlier genes e.g., SMT3H1
- RNA processing e.g., RDBP
- metabolic processes e.g., NME1
- outlier genes e.g., SMT3H1
- SMT3H1 are members of the ubiquitin-proteasome pathway, suggesting deregulation of this pathway in HCC.
- a gene cluster associated with lymphocyte infiltrate that included the expression of genes such as IGKC and IGJ was observed, and transcription factors (e.g.
- ERBB3 is defective in tyrosine kinase activity and requires dimerization with other receptors, possibly another member of the ERBB family (Riese and Stern, 1998), the hypothesis that HCC tumors expressing high levels of ERBB3 were associated with high expression of ERRB2 or EGFR was tested.
- the expression of ERBB2 was elevated in 12 of 37 tumors ( Figure 5C), while high EGFR expression was found in 15 of 37 tumors ( Figure 5D).
- a cholesterol biosynthetic pathway was characterized by higher expression in HCC tumors for genes.of the enzymes SQLE, ACLY and FDPS.
- a subgroup involved in growth and differentiation was characterized in the HCC tumor tissues by lower expression of ESRl, IGFBP3 and PDGFR ⁇ , and high expression of PPTB1.
- a subgroup of bZIP transcription factors ATF3, FOS, JUN, and MYBL2 was characterized to be down-regulated in the HCC tumor tissues.
- the discriminator cassettes were assessed on an independent tissue set of 58 liver clinical biopsies from 29 patients. Using a kNN prediction algorithm, it was found that all classifier probe cassettes could readily distinguish HCC tumor from non-tumor liver (Table 5), and that the gene discriminators of tumor vs. non-tumor in HCC derived by the intersect analysis of limited tissue sets can be validated in an independent manner. Table 5. Prediction accuracy of gene classifiers using k NN algorithm on 58 liver biopsies from 29 patients.
- False negative cases refer to HCC tumors which were misclassified as non-tumor livers. *False positive cases refer to non-tumor livers which were misclassified as HCC tumors.
- the anaphase-promoting complex is required in Gl arrested yeast cells to inhibit B-tyoe cyclin accumulation and to prevent uncontrolled entry into S-phase. J. Cell Sci. 110, 1523-1531.
- Bmi-1 determines the proliferative capacity of normal and leukaemic stem cells. Nature 423, 255-260. Leung, T. H. Y., Wong, N., Lai, P. B. S., Chan, A., To, K-F., Liew, C. T., Lau, W-Y., and Johnson, P. J. (2002). Identification of four distinct regions of allelic imbalances on chromosome 1 by the combined comparative genomic hybridization and microsatellite analysis on hepatocellular carcinoma. Mod. Pathol. 15, 1213-1220.
- Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423, 302-305. Parkin, D. M., Pisani, P., and Ferlay, J. (1999). Global cancer statistics. CA Cancer J. Clin. 49, 33-64.
- Core biopsies can be used to distinguish differences in expression profiling by cDNA microarrays. J. Mol. Diag. 4, 30-36.
- the polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419, 624-629.
- the bmi-1 oncoprotein is differentially expressed in non-small cell lung cancer and correlates with INK4A-ARF locus expression.
- Pleitrophin and midkine a family of mitogenic and angiogenic heparin-binding growth and differentiation factors. Curr. Opin. Hematol. 6, 44-50.
- Accession numbers may refer to Accession numbers from multiple databases, including GenBank, the European Molecular Biology Laboratory (EMBL), the DNA Database of Japan (DDBJ), or the Genome Sequence Data Base (GSDB), for nucleotide sequences, and including the Protein Information Resource (PIR), SWISSPROT, Protein Research Foundation (PRF), and Protein Data Bank (PDB) (sequences from solved structures), as well as from translations from annotated coding regions from nucleotide sequences in GenBank, EMBL, DDBJ, or RefSeq, for polypeptide sequences. Accession numbers, as used herein, may also refer to Accession numbers from databases such as UniGene, OMIM, LocusLink, or HomoloGene.
Landscapes
- Health & Medical Sciences (AREA)
- Chemical & Material Sciences (AREA)
- Life Sciences & Earth Sciences (AREA)
- Organic Chemistry (AREA)
- General Health & Medical Sciences (AREA)
- Zoology (AREA)
- Medicinal Chemistry (AREA)
- Genetics & Genomics (AREA)
- Engineering & Computer Science (AREA)
- Proteomics, Peptides & Aminoacids (AREA)
- Bioinformatics & Cheminformatics (AREA)
- Biochemistry (AREA)
- Wood Science & Technology (AREA)
- Molecular Biology (AREA)
- Immunology (AREA)
- Pharmacology & Pharmacy (AREA)
- Animal Behavior & Ethology (AREA)
- Chemical Kinetics & Catalysis (AREA)
- Nuclear Medicine, Radiotherapy & Molecular Imaging (AREA)
- Biophysics (AREA)
- Veterinary Medicine (AREA)
- General Chemical & Material Sciences (AREA)
- Public Health (AREA)
- Gastroenterology & Hepatology (AREA)
- Toxicology (AREA)
- Pathology (AREA)
- Microbiology (AREA)
- Biotechnology (AREA)
- General Engineering & Computer Science (AREA)
- Analytical Chemistry (AREA)
- Hospice & Palliative Care (AREA)
- Oncology (AREA)
- Biomedical Technology (AREA)
- Physics & Mathematics (AREA)
- Measuring Or Testing Involving Enzymes Or Micro-Organisms (AREA)
- Apparatus Associated With Microorganisms And Enzymes (AREA)
- Medicines That Contain Protein Lipid Enzymes And Other Medicines (AREA)
- Investigating Or Analysing Biological Materials (AREA)
Abstract
Description
Claims
Priority Applications (2)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| EP04736172A EP1631682A1 (en) | 2003-06-04 | 2004-06-04 | Differentially regulated hepatocellular carcinoma genes and uses thereof |
| JP2006508583A JP2006526405A (en) | 2003-06-04 | 2004-06-04 | Differentially regulated hepatocellular oncogene and its utilization |
Applications Claiming Priority (2)
| Application Number | Priority Date | Filing Date | Title |
|---|---|---|---|
| US47550803P | 2003-06-04 | 2003-06-04 | |
| US60/475,508 | 2003-06-04 |
Publications (1)
| Publication Number | Publication Date |
|---|---|
| WO2004108964A1 true WO2004108964A1 (en) | 2004-12-16 |
Family
ID=33511687
Family Applications (1)
| Application Number | Title | Priority Date | Filing Date |
|---|---|---|---|
| PCT/SG2004/000166 WO2004108964A1 (en) | 2003-06-04 | 2004-06-04 | Differentially regulated hepatocellular carcinoma genes and uses thereof |
Country Status (3)
| Country | Link |
|---|---|
| EP (1) | EP1631682A1 (en) |
| JP (1) | JP2006526405A (en) |
| WO (1) | WO2004108964A1 (en) |
Cited By (15)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| JP2007006800A (en) * | 2005-06-30 | 2007-01-18 | Japan Science & Technology Agency | Normal human hepatocyte-specific genes |
| WO2008102002A3 (en) * | 2007-02-21 | 2008-11-27 | Rikshospitalet Radiumhospitale | New markers for cancer |
| JP2010508811A (en) * | 2006-11-07 | 2010-03-25 | チョンブク ナショナル ユニバーシティー インダストリアル コーポレーション ファンデーション | Diagnostic composition for hepatocellular carcinoma, diagnostic kit for hepatocellular carcinoma comprising the same, and diagnostic method for hepatocellular carcinoma |
| EP2187216A1 (en) * | 2007-08-10 | 2010-05-19 | Hyogo College Of Medicine | Novel liver cancer marker |
| WO2010072383A1 (en) * | 2008-12-22 | 2010-07-01 | 7Roche Diagnostics Gmbh | Armet as a marker for cancer |
| US20110172288A1 (en) * | 2007-01-24 | 2011-07-14 | Dana-Farber Cancer Institute, Inc. | Compositions and Methods for the Identification, Assessment, Prevention and Therapy of Thymic Lymphoma or Hamartomatous Tumours |
| US8030013B2 (en) * | 2006-04-14 | 2011-10-04 | Mount Sinai School Of Medicine | Methods and compositions for the diagnosis for early hepatocellular carcinoma |
| EP2574679A3 (en) * | 2005-11-30 | 2013-07-31 | Institut National De La Sante Et De La Recherche Medicale (Inserm) | Methods for in vitro hepatocellular carninoma prognosis |
| US20140171335A1 (en) * | 2007-03-26 | 2014-06-19 | Novartis Ag | Predictive renal safety biomarkers and biomarker signatures to monitor kidney function |
| WO2015093932A1 (en) * | 2013-12-18 | 2015-06-25 | Instituto Nacional De Medicina Genómica | Method for the early diagnosis of hepatocellular carcinoma |
| WO2016056995A1 (en) * | 2014-10-09 | 2016-04-14 | Singapore Health Services Pte Ltd | Profiling and/or therapy of hepatocellular carcinoma |
| US9382318B2 (en) | 2012-05-18 | 2016-07-05 | Amgen Inc. | ST2 antigen binding proteins |
| CN110218788A (en) * | 2018-03-02 | 2019-09-10 | 香港中文大学 | Application of the squalene epoxidase in the diagnosing and treating of non-alcohol fatty liver |
| WO2020189525A1 (en) | 2019-03-15 | 2020-09-24 | 日本曹達株式会社 | Heterocyclic compound and agricultural or horticultural bactericide |
| AU2020244420B2 (en) * | 2015-05-06 | 2022-08-25 | Immatics Biotechnologies Gmbh | Novel peptides and combination of peptides and scaffolds thereof for use in immunotherapy against colorectal carcinoma (crc) and other cancers |
Families Citing this family (3)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| WO2010048304A2 (en) * | 2008-10-21 | 2010-04-29 | Bayer Healthcare Llc | Identification of signature genes associated with hepatocellular carcinoma |
| JP2014027898A (en) * | 2012-07-31 | 2014-02-13 | Yamaguchi Univ | Method for judging onset risk of hepatocarcinoma |
| CN118166038B (en) * | 2024-05-10 | 2024-08-09 | 广州明迅生物科技有限责任公司 | Method for constructing immunodeficiency animal model |
-
2004
- 2004-06-04 WO PCT/SG2004/000166 patent/WO2004108964A1/en not_active Application Discontinuation
- 2004-06-04 EP EP04736172A patent/EP1631682A1/en not_active Withdrawn
- 2004-06-04 JP JP2006508583A patent/JP2006526405A/en not_active Ceased
Non-Patent Citations (8)
Cited By (25)
| Publication number | Priority date | Publication date | Assignee | Title |
|---|---|---|---|---|
| JP2007006800A (en) * | 2005-06-30 | 2007-01-18 | Japan Science & Technology Agency | Normal human hepatocyte-specific genes |
| EP2574679A3 (en) * | 2005-11-30 | 2013-07-31 | Institut National De La Sante Et De La Recherche Medicale (Inserm) | Methods for in vitro hepatocellular carninoma prognosis |
| US8030013B2 (en) * | 2006-04-14 | 2011-10-04 | Mount Sinai School Of Medicine | Methods and compositions for the diagnosis for early hepatocellular carcinoma |
| JP2010508811A (en) * | 2006-11-07 | 2010-03-25 | チョンブク ナショナル ユニバーシティー インダストリアル コーポレーション ファンデーション | Diagnostic composition for hepatocellular carcinoma, diagnostic kit for hepatocellular carcinoma comprising the same, and diagnostic method for hepatocellular carcinoma |
| US9632090B2 (en) * | 2007-01-24 | 2017-04-25 | Dana-Farber Cancer Institute, Inc. | Compositions and methods for the identification, assessment, prevention and therapy of thymic lymphoma or hamartomatous tumours |
| US20110172288A1 (en) * | 2007-01-24 | 2011-07-14 | Dana-Farber Cancer Institute, Inc. | Compositions and Methods for the Identification, Assessment, Prevention and Therapy of Thymic Lymphoma or Hamartomatous Tumours |
| EP2474628A1 (en) * | 2007-02-21 | 2012-07-11 | Oslo Universitetssykehus HF | New markers for cancer |
| WO2008102002A3 (en) * | 2007-02-21 | 2008-11-27 | Rikshospitalet Radiumhospitale | New markers for cancer |
| US20140171335A1 (en) * | 2007-03-26 | 2014-06-19 | Novartis Ag | Predictive renal safety biomarkers and biomarker signatures to monitor kidney function |
| EP2187216A1 (en) * | 2007-08-10 | 2010-05-19 | Hyogo College Of Medicine | Novel liver cancer marker |
| CN101796415B (en) * | 2007-08-10 | 2014-03-12 | 学校法人兵库医科大学 | Novel liver cancer marker |
| EP2187216A4 (en) * | 2007-08-10 | 2011-03-02 | Hyogo College Medicine | Novel liver cancer marker |
| US8741587B2 (en) | 2008-12-22 | 2014-06-03 | Roche Diagnostics Operations, Inc. | ARMET as a marker for cancer |
| WO2010072383A1 (en) * | 2008-12-22 | 2010-07-01 | 7Roche Diagnostics Gmbh | Armet as a marker for cancer |
| US11059895B2 (en) | 2012-05-18 | 2021-07-13 | Amgen Inc. | ST2 antigen binding proteins |
| US9382318B2 (en) | 2012-05-18 | 2016-07-05 | Amgen Inc. | ST2 antigen binding proteins |
| US9982054B2 (en) | 2012-05-18 | 2018-05-29 | Amgen Inc. | ST2 antigen binding proteins |
| US10227414B2 (en) | 2012-05-18 | 2019-03-12 | Amgen Inc. | ST2 antigen binding proteins |
| US11965029B2 (en) | 2012-05-18 | 2024-04-23 | Amgen Inc. | ST2 antigen binding proteins |
| WO2015093932A1 (en) * | 2013-12-18 | 2015-06-25 | Instituto Nacional De Medicina Genómica | Method for the early diagnosis of hepatocellular carcinoma |
| WO2016056995A1 (en) * | 2014-10-09 | 2016-04-14 | Singapore Health Services Pte Ltd | Profiling and/or therapy of hepatocellular carcinoma |
| AU2020244420B2 (en) * | 2015-05-06 | 2022-08-25 | Immatics Biotechnologies Gmbh | Novel peptides and combination of peptides and scaffolds thereof for use in immunotherapy against colorectal carcinoma (crc) and other cancers |
| CN110218788A (en) * | 2018-03-02 | 2019-09-10 | 香港中文大学 | Application of the squalene epoxidase in the diagnosing and treating of non-alcohol fatty liver |
| WO2020189525A1 (en) | 2019-03-15 | 2020-09-24 | 日本曹達株式会社 | Heterocyclic compound and agricultural or horticultural bactericide |
| KR20210138585A (en) | 2019-03-15 | 2021-11-19 | 닛뽕소다 가부시키가이샤 | Heterocyclic compounds and agricultural and horticultural fungicides |
Also Published As
| Publication number | Publication date |
|---|---|
| EP1631682A1 (en) | 2006-03-08 |
| JP2006526405A (en) | 2006-11-24 |
Similar Documents
| Publication | Publication Date | Title |
|---|---|---|
| JP4938672B2 (en) | Methods, systems, and arrays for classifying cancer, predicting prognosis, and diagnosing based on association between p53 status and gene expression profile | |
| Neo et al. | Identification of discriminators of hepatoma by gene expression profiling using a minimal dataset approach | |
| Woo et al. | Association of TP53 mutations with stem cell-like gene expression and survival of patients with hepatocellular carcinoma | |
| WO2004108964A1 (en) | Differentially regulated hepatocellular carcinoma genes and uses thereof | |
| US8722331B2 (en) | Method for selecting a treatment for non-small cell lung cancer using gene expression profiles | |
| US20040146921A1 (en) | Expression profiles for colon cancer and methods of use | |
| Pressey et al. | Hedgehog pathway activity in pediatric embryonal rhabdomyosarcoma and undifferentiated sarcoma: a report from the Children's Oncology Group | |
| EP2253721A1 (en) | Method for predicting the occurence of lung metastasis in breast cancer patients | |
| EP1945819B1 (en) | Gene expression profiles and methods of use | |
| US20070015148A1 (en) | Gene expression profiles in breast tissue | |
| WO2013088457A1 (en) | Genetic variants useful for risk assessment of thyroid cancer | |
| US20070065827A1 (en) | Gene expression profiles and methods of use | |
| US20140031242A1 (en) | Method for discovering pharmacogenomic biomarkers | |
| US20060240441A1 (en) | Gene expression profiles and methods of use | |
| EP1605811A2 (en) | Expression profiles for breast cancer and methods of use | |
| US20160201143A1 (en) | LKB1 Levels and Brain Metastasis from Non-Small-Cell Lung Cancer (NSCLC) | |
| WO2011153345A2 (en) | A gene expression profile of brca-ness that correlates with responsiveness to chemotherapy and with outcome in cancer patients | |
| US20150284806A1 (en) | Materials and methods for determining susceptibility or predisposition to cancer | |
| WO2011056963A1 (en) | Methods and compositions for predicting survival in subjects with cancer | |
| US20120129711A1 (en) | Biomarkers for the prognosis and high-grade glioma clinical outcome | |
| WO2017079695A9 (en) | Gene expression patterns to predict responsiveness to virotherapy in cancer indications | |
| US20050147970A1 (en) | Breast cancer associated polypeptide | |
| Furge et al. | Gene expression profiling in kidney cancer: combining differential expression and chromosomal and pathway analyses | |
| Mingjian et al. | Genetic fingerprint concerned with lymphatic metastasis of human lung squamous cancer | |
| CN118866075A (en) | System for detecting liver cancer |
Legal Events
| Date | Code | Title | Description |
|---|---|---|---|
| AK | Designated states |
Kind code of ref document: A1 Designated state(s): AE AG AL AM AT AU AZ BA BB BG BR BW BY BZ CA CH CN CO CR CU CZ DE DK DM DZ EC EE EG ES FI GB GD GE GH GM HR HU ID IL IN IS JP KE KG KP KR KZ LC LK LR LS LT LU LV MA MD MG MK MN MW MX MZ NA NI NO NZ OM PG PH PL PT RO RU SC SD SE SG SK SL SY TJ TM TN TR TT TZ UA UG US UZ VC VN YU ZA ZM ZW |
|
| AL | Designated countries for regional patents |
Kind code of ref document: A1 Designated state(s): BW GH GM KE LS MW MZ NA SD SL SZ TZ UG ZM ZW AM AZ BY KG KZ MD RU TJ TM AT BE BG CH CY CZ DE DK EE ES FI FR GB GR HU IE IT LU MC NL PL PT RO SE SI SK TR BF BJ CF CG CI CM GA GN GQ GW ML MR NE SN TD TG |
|
| 121 | Ep: the epo has been informed by wipo that ep was designated in this application | ||
| WWE | Wipo information: entry into national phase |
Ref document number: 2006508583 Country of ref document: JP |
|
| WWE | Wipo information: entry into national phase |
Ref document number: 2004736172 Country of ref document: EP |
|
| WWW | Wipo information: withdrawn in national office |
Ref document number: 2004736172 Country of ref document: EP |
|
| WWP | Wipo information: published in national office |
Ref document number: 2004736172 Country of ref document: EP |