DART: Deformable Anatomy-aware Registration Toolkit for lung ct registration with keypoints supervision
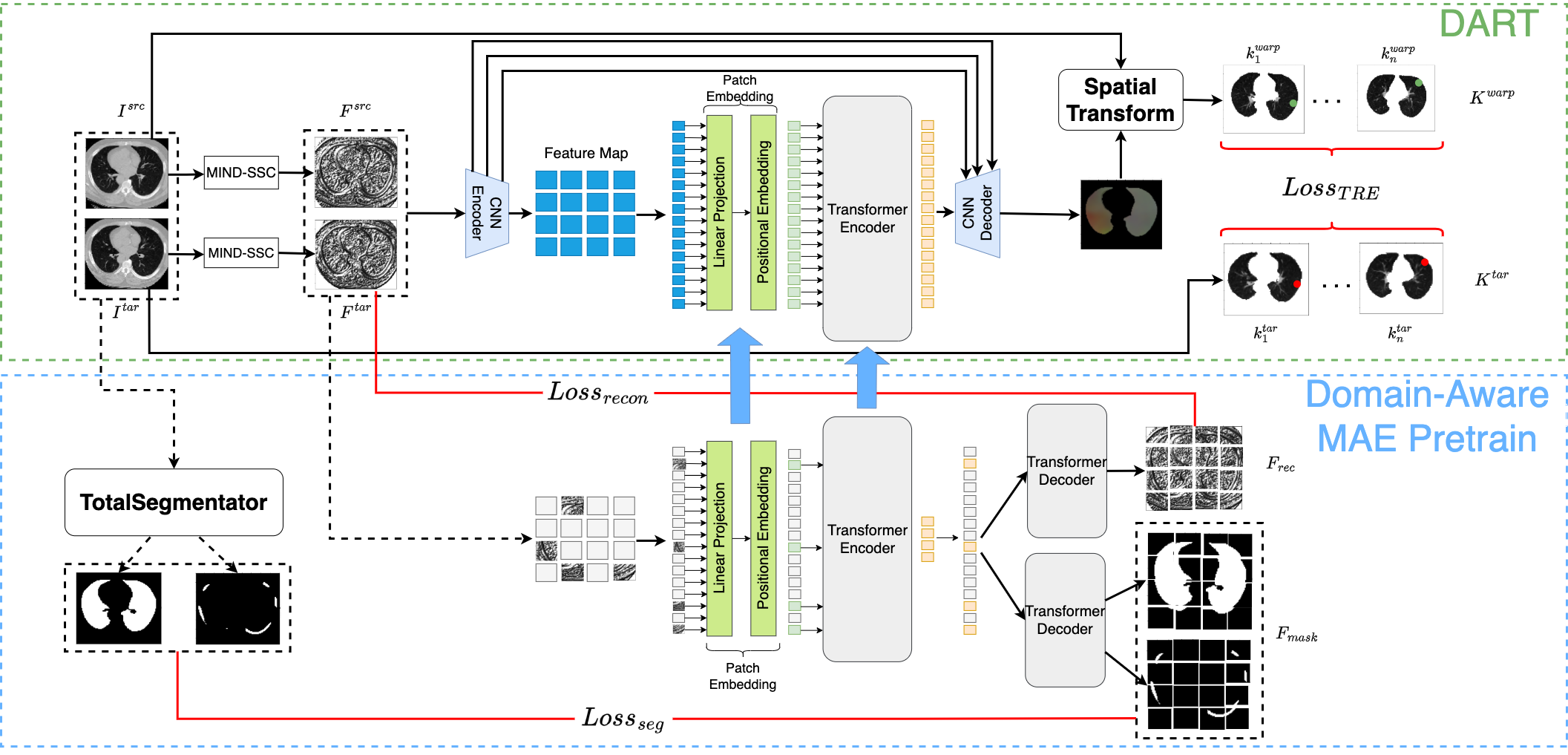
Figure 1. An overview of the proposed DART.
Spatially aligning two computed tomography (CT) scans of the lung using automated image registration techniques is a challenging task due to the deformable nature of the lung. However, existing deep-learning-based lung CT registration models are trained with no prior knowledge of anatomical understanding. We propose the deformable anatomy-aware registration toolkit (DART), a masked autoencoder (MAE)-based approach, to improve the keypoint-supervised registration of lung CTs. Our method incorporates features from multiple decoders of networks trained to segment anatomical structures, including the lung, ribs, vertebrae, lobes, vessels, and airways, to ensure that the MAE learns relevant features corresponding to the anatomy of the lung. The pretrained weights of the transformer encoder and patch embeddings are then used as the initialization for the training of downstream registration. We compare DART to existing state-of the-art registration models. Our experiments show that DART outperforms the baseline models (Voxelmorph, ViT-V-Net, and MAE-TransRNet) in terms of target registration error of both corrField-generated keypoints with 17%, 13%, and 9% relative improvement, respectively, and bounding box centers of nodules with 27%, 10%, and 4% relative improvement, respectively.
git clone https://github.com/yunzhengzhu/DART.git
cd DARTPlease download from link and save the weights folder under your DART folder.
- NVIDIA GPU
- python 3.8.12
- pytorch 1.11.0a0+b6df043
- numpy 1.24.2
- pandas 1.3.4
- nibabel 5.1.0
- scipy 1.9.1
- natsort 8.4.0
- einops 0.7.0
- matplotlib 3.5.0
- tensorboardX 2.6.2.2
- scikit-learn 0.24.0
- torchio 0.19.6
Install pytorch and python. You can install other dependencies by
pip install -r requirements.txtDownload NLST from Learn2Reg Datasets. Note: getting access requires registering an account and joining the challenge. The entire dataset consists of 210 pairs of lung CT scans (fixed as baseline and moving as followup) is already preprocessed to a fixed sized 224 x 192 x 224 with spacing 1.5 x 1.5 x 1.5. The corresponding lung masks and keypoints are also provided. For details, please refer to Learn2Reg2023.
cd DATA
unzip NLST2023.zipPlease refer to TotalSegmentator for generating the masks (lung, lung lobes, pulmonary vessels, airways, vertebrates, ribs, etc.)
Note: Please flipping the image to RAS orientation before using totalsegmentator, and flipping the segmentation back to the same orientation as the registration dataset.
Please refer to MONAI lung nodule detection for generating the stats (box coordinates and probabilities) for lung nodule bounding boxes. The centers of the bounding box are used to evaluate the registration as the TRE_nodule metric.
Training with baselines require no pretraining (Voxelmorph, ViT-V-Net)
CUDA_VISIBLE_DEVICES='0' main.py --data_dir DATA/NLST2023 --json_file NLST_dataset.json --result_dir exp --exp_name test --mind_feature --preprocess --use_scaler --downsample 2 --model_type 'Vxm' --loss 'TRE' --loss_weight 1.0 --diff --opt 'adam' --lr 1e-4 --sche 'lambdacosine' --max_epoch 300.0 --lrf 0.01 --batch_size 1 --epochs 300 --seed 1234 --es --es_warmup 0 --es_patience 300 --es_criterion 'TRE' --log --print_every 10Note: For model_type in downstream registration, you need to modify the argument for different models (Vxm for Voxelmorph, ViT-V-Net for ViT-V-Net, MAE-ViT_Baseline for MAE-TransRNet, MAE_ViT_Seg for DART)
Note: You could skip this step by using our trained weights from weights/registration/vxm/es_checkpoint.pth.tar or weights/registration/vitvnet/es_checkpoint.pth.tar
exp_dir=weights/registration/vxm
CUDA_VISIBLE_DEVICES='0' eval.py --exp_dir ${exp_dir} --save_df --save_warped --eval_diff --mode valA results_val.csv will be generated under your ${exp_dir} folder.
Training with baselines require pretraining (MAE-TransRNet)
CUDA_VISIBLE_DEVICES='0' pretrain_baseline.py --data_dir DATA/NLST2023 --json_file NLST_dataset.json --result_dir exp --exp_name pt_test --preprocess --use_scaler --mind_feature --downsample 8 --model_type 'MAE_ViT' --loss 'MSE' --loss_weight 1.0 --opt 'adam' --lr 1e-4 --sche 'lambdacosine' --max_epoch 300.0 --lrf 0.01 --batch_size 1 --epochs 1 --seed 1234 --es --es_warmup 0 --es_patience 300 --es_criterion 'MSE' --log --print_every 10Note: You could skip this step by using our trained weights from weights/pretraining/mae_t.pth.tar
Same script as Downstream Registration above, but remember to load the pretrained weights by adding the argument --pretrained ${pretrained_weights}. Remember to specify model_type as MAE_ViT_Baseline before running.
Note: You could skip this step by using our trained weights from weights/registration/mae_t/es_checkpoint.pth.tar
exp_dir=weights/registration/mae_t
CUDA_VISIBLE_DEVICES='0' eval.py --exp_dir ${exp_dir} --save_df --save_warped --eval_diff --mode valA results_val.csv will be generated under your ${exp_dir} folder.
Note: Please prepare segmentation masks (Lung, Lung Lobes, Airways, Pulmonary Vessels, etc.) before doing the following steps.
mask_dir=DATA/TOTALSEG_MASK
CUDA_VISIBLE_DEVICES='0' pretrain_baseline.py --data_dir DATA/NLST2023 --json_file NLST_dataset.json --result_dir exp --exp_name pt_test --preprocess --use_scaler --mind_feature --downsample 8 --model_type 'MAE_ViT_Seg' --loss 'MSE' 'Seg_MSE' --loss_weight 1.0 1.0 --opt 'adam' --lr 1e-4 --sche 'lambdacosine' --max_epoch 300.0 --lrf 0.01 --batch_size 1 --epochs 1 --seed 1234 --es --es_warmup 0 --es_patience 300 --es_criterion 'MSE' --log --print_every 10 --mask_dir ${mask_dir} --eval_with_mask --specific_regions 'lung_trachea_bronchia'Note: Arguments particularly designed for DART: (examples are using filenames from TotalSegmentator)
mask_dir: specify the dir saving the generated masks
eval_with_mask: using the generated masks or not
specific_regions: specific regions used for anatomy-aware pretraining
- airways (A):
'lung_trachea_bronchia' - vessels (Ve.):
'lung_vessels' - 5 lobes (Lo.):
'lung_lower_lobe_left' 'lung_lower_lobe_right' 'lung_middle_lobe_right' 'lung_upper_lobe_left' 'lung_upper_lobe_right'
organs: specify the organ (files contain this organ's name)
- lung + rib (LR):
'lung' 'rib' - lung + vertebrae (LV):
'lung' 'vertebrae' - lung + rib + vertebrae (LRV):
'lung' 'rib' 'vertebrae'
Note: You could skip this step by using our trained weights from weights/pretraining/dart_airways.pth.tar
Same script as Downstream Registration above, but remember to load the pretrained weights by adding the argument --pretrained weights/dart_airways.pth.tar or your own trained weights. Remember to specify model_type as MAE_ViT_Baseline before running.
Note: You could skip this step by using our trained weights from weights/registration/dart_airways/es_checkpoint.pth.tar
exp_dir=weights/registration/dart_airways
CUDA_VISIBLE_DEVICES='0' eval.py --exp_dir ${exp_dir} --save_df --save_warped --eval_diff --mode valA results_val.csv will be generated under your ${exp_dir} folder.
TRE_kp: Target registration error of corrField-generated keypoints
TRE_nodule: Target registration error of bounding box centers generated from MONAI nodule detection algorithm (Please see Nodule Center Generation section above)
SDLogJ: std of the logarithm of the Jacobian determinant of the displacement vector field