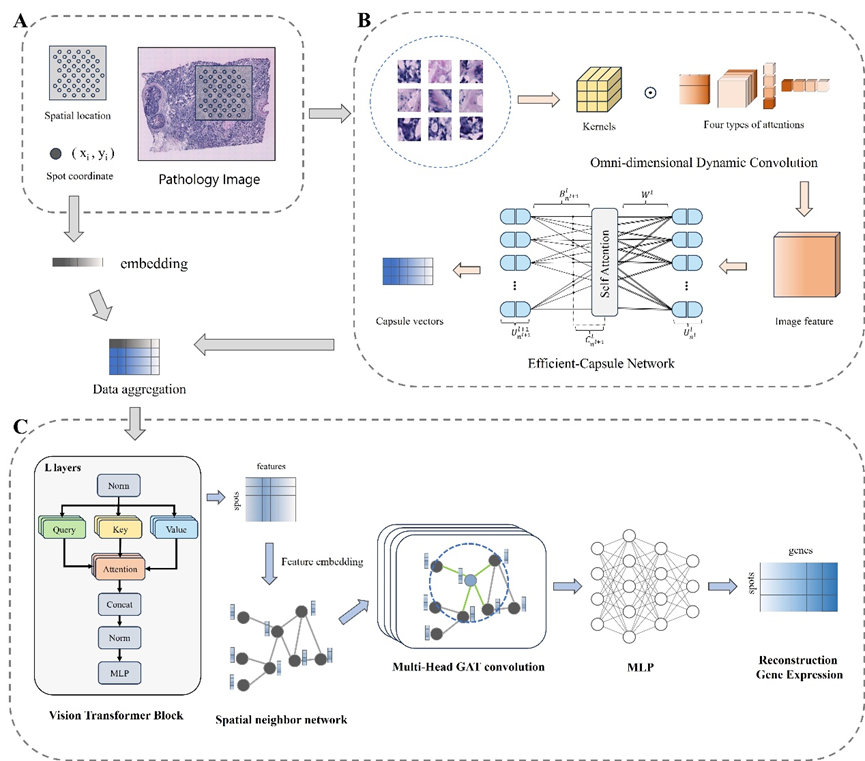
THItoGene is a hybrid neural network that leverages dynamic convolution and capsule networks to adaptively perceive
latent molecular signals from histological images, for the systematic analysis of spatial gene expression within tissue
pathology. THItoGene integrates gene expression, spatial locations, and histological images to explore and analyze the
relationship between high-resolution pathological image phenotypes and tumor genetic morphology.
The required environment has been packaged in the requirements.txt file.
Please run the following command to install.
cd THItoGene
pip install -r requirements.txt
- Human HER2-positive breast tumor ST data https://github.com/almaan/her2st/.
- Human cutaneous squamous cell carcinoma 10x Visium data (GSE144240).
- You can also download all datasets from here
All Trained models of our method on HER2+ and cSCC datasets can be found at synapse.
NOTE: Please download our trained models and datasets first and extract them to the corresponding folder.
import torch
from torch.utils.data import DataLoader
from dataset import ViT_HER2ST
from predict import model_predict
from utils import *
from vis_model import THItoGene
test_sample_ID = 0
dataset = 'her2st'
# Model loading(Please unzip the trained model into the model folder first)
model = THItoGene.load_from_checkpoint(
f"model/THItoGene_{dataset}_{test_sample_ID}.ckpt", n_genes=785,
learning_rate=1e-5, route_dim=64, caps=20, heads=[16, 8],
n_layers=4)
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
# Data set loading
dataset = ViT_HER2ST(train=False, sr=False, fold=test_sample_ID)
test_loader = DataLoader(dataset, batch_size=1, num_workers=0)
# Model prediction
adata_pred, adata_truth = model_predict(model, test_loader, attention=False, device=device)
# Evaluation
R, p_val = get_R(adata_pred, adata_truth)
print('Mean Pearson Correlation:', np.nanmean(R))
print('-log10p_val:', -np.log10(p_val))n_genes: int.
Amount of genes.learning_rate: float between[0, 1], default1e-5.
Learning rate.route_dim: int, default64.
Capsule network routing vector dimension.heads: int, default[16, 8].
The number of heads of the Vit module and the number of heads of the GAT module.n_layers: int, default4.
Number of Transformer blocks.caps: int, default20.
Capsule network routing capsule number.
NOTE: Run the following command if you want to run the pipline
-
Please run the script
download.shin the folder data (or run the command linegit clone https://github.com/almaan/her2st.gitin the dir data) -
Run
gunzip *.gzin the dir./data/her2st/data/ST-cnts/to unzip the gz files
import pytorch_lightning as pl
from torch.utils.data import DataLoader
from dataset import ViT_HER2ST
from vis_model import THItoGene
fold = 0
tag = '-htg_her2st_785_32_cv'
dataset = ViT_HER2ST(train=True, fold=fold)
train_loader = DataLoader(dataset, batch_size=1, num_workers=0, shuffle=True)
model = THItoGene(n_genes=785, learning_rate=1e-5, route_dim=64, caps=20, heads=[16, 8], n_layers=4)
trainer = pl.Trainer(accelerator="gpu", devices=[0], max_epochs=200)
trainer.fit(model, train_loader)
trainer.save_checkpoint("model/last_train_" + tag + '_' + str(fold) + ".ckpt")import torch
from torch.utils.data import DataLoader
from dataset import ViT_HER2ST
from predict import model_predict
from utils import *
from vis_model import THItoGene
fold = 0
tag = '-htg_her2st_785_32_cv'
model = THItoGene.load_from_checkpoint("model/last_train_" + tag + '_' + str(fold) + ".ckpt", n_genes=785,
learning_rate=1e-5, route_dim=64, caps=20, heads=[16, 8],
n_layers=4)
device = torch.device('cuda' if torch.cuda.is_available() else 'cpu')
dataset = ViT_HER2ST(train=False, sr=False, fold=fold)
test_loader = DataLoader(dataset, batch_size=1, num_workers=4)
adata_pred, adata_truth = model_predict(model, test_loader, attention=False, device=device)
R, p_val = get_R(adata_pred, adata_truth)
print('Mean Pearson Correlation:', np.nanmean(R))
print('-log10p_val:', -np.log10(p_val))Jia et al. “THItoGene: a deep learning method for predicting spatial transcriptomics from histological images.” Briefings in bioinformatics vol. 25,1 (2024).Paper.