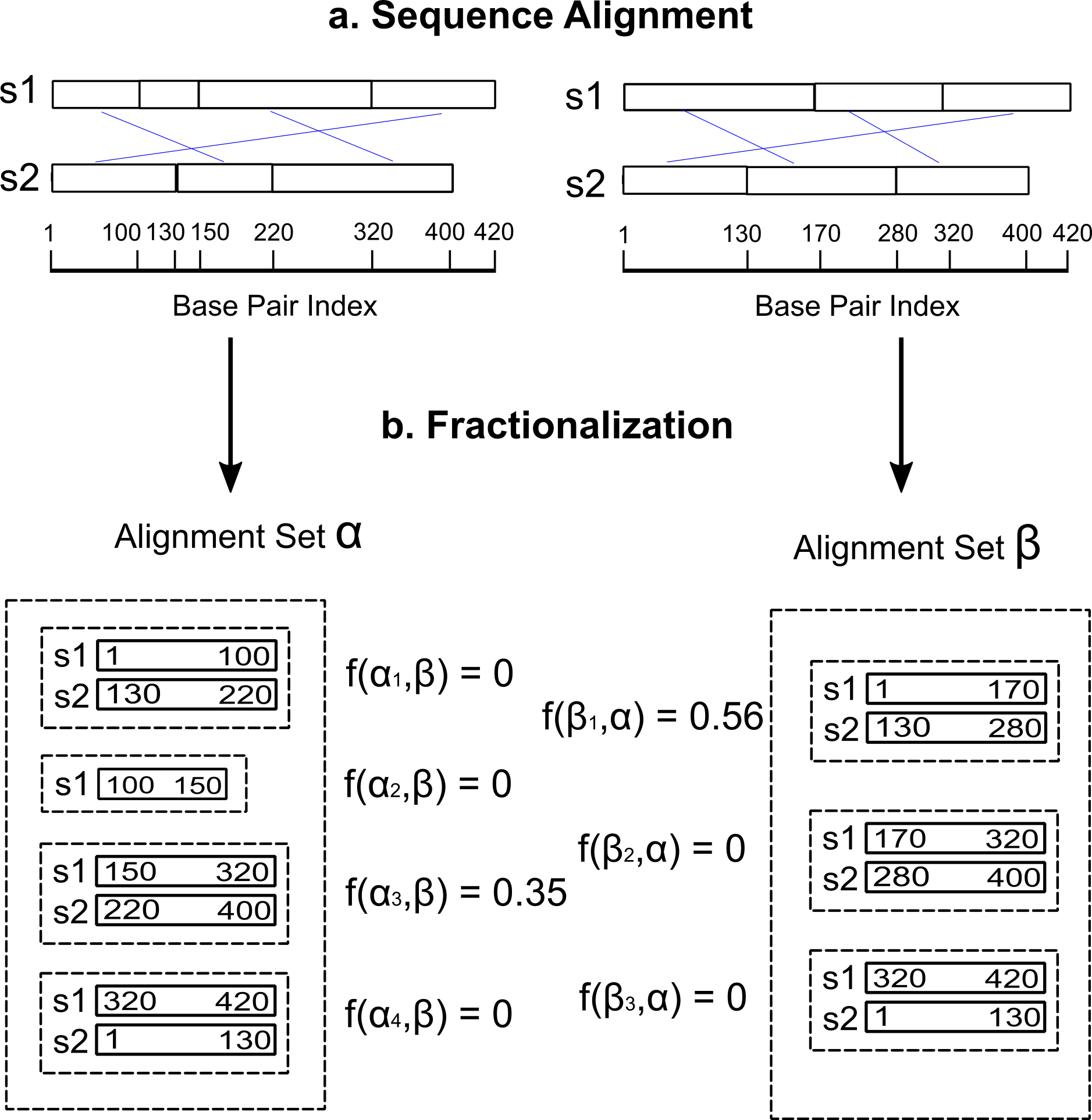
Fractionalization is a value commonly used in socioeconomics measuring how a society can be divived into different groups accoding to a given dimension such as a race, a religion, or a language. We, taking the concept of fractionalization and applying it into genome alignment comparison, implemented this script which takes two whole genome alignment files as input, and outputs the fractionalization values which quantify how congruent the two alignment files are.
A fractionalization value is generated for each multiple sequence alignment, calculated as
where
Now, the script supports maf, mhg, xmfa file format generated by Cactus, Mauve, MHG/MHG-EVO, and SibeliaZ. For cactus output, which is in hal format, in order to calculate the fractionalization value set, please convert the hal output to maf format using hal2maf provided in Cactus. For the output, this script will output a tsv file for each of the input alignment file containing the length of each alignment, the number of sequence in each alignment, and the fractionalization value corresponding to the alignment comparing with the other alignment file.
usage: frac_calculate [-h] -g GENOME -a ALIGNMENTA -b ALIGNMENTB -at {mhg,mauve,cactus,sibeliaz} -bt
{mhg,mauve,cactus,sibeliaz} [-gb GENOME_BED_OUTPUT] [-pa PREFIXA] [-pb PREFIXB] [-t THRESHOLD]
Fractionalization Calculation to Compare Two Whole Genome Alignment Results
optional arguments:
-h, --help show this help message and exit
-g GENOME, --genome GENOME
A directory of input fasta genomes for whole genome aligners, will read in all files ended
with '.fna', '.fa', '.fasta'
-a ALIGNMENTA, --alignmentA ALIGNMENTA
Path to the 1st alignment file
-b ALIGNMENTB, --alignmentB ALIGNMENTB
Path to the 2nd alignment file
-at {mhg,mauve,cactus,sibeliaz}, --AType {mhg,mauve,cactus,sibeliaz}
File format of the 1st alignment file; 'txt' for mhg/mhg-evo; 'xmfa' for mauve; 'maf' for
cactus/sibeliaz
-bt {mhg,mauve,cactus,sibeliaz}, --BType {mhg,mauve,cactus,sibeliaz}
File format of the 2nd alignment file; 'txt' for mhg/mhg-evo; 'xmfa' for mauve; 'maf' for
cactus/sibeliaz
-gb GENOME_BED_OUTPUT, --genome_bed_output GENOME_BED_OUTPUT
Output path for genome bed file recording all contig length, default to 'genome.bed'
-pa PREFIXA, --prefixA PREFIXA
Prefix for the 1st alignment file, default is 'A'
-pb PREFIXB, --prefixB PREFIXB
Prefix for the 2nd alignment file, default is 'B'
-t THRESHOLD, --threshold THRESHOLD
If an alignment is shorter than the threshold, its fractionalization value will not be
reported to avoid an over-representation for the total fractionalization values being too high
or too low. If it is needed to consider all alignments no matter for those being short, set
this value to 0