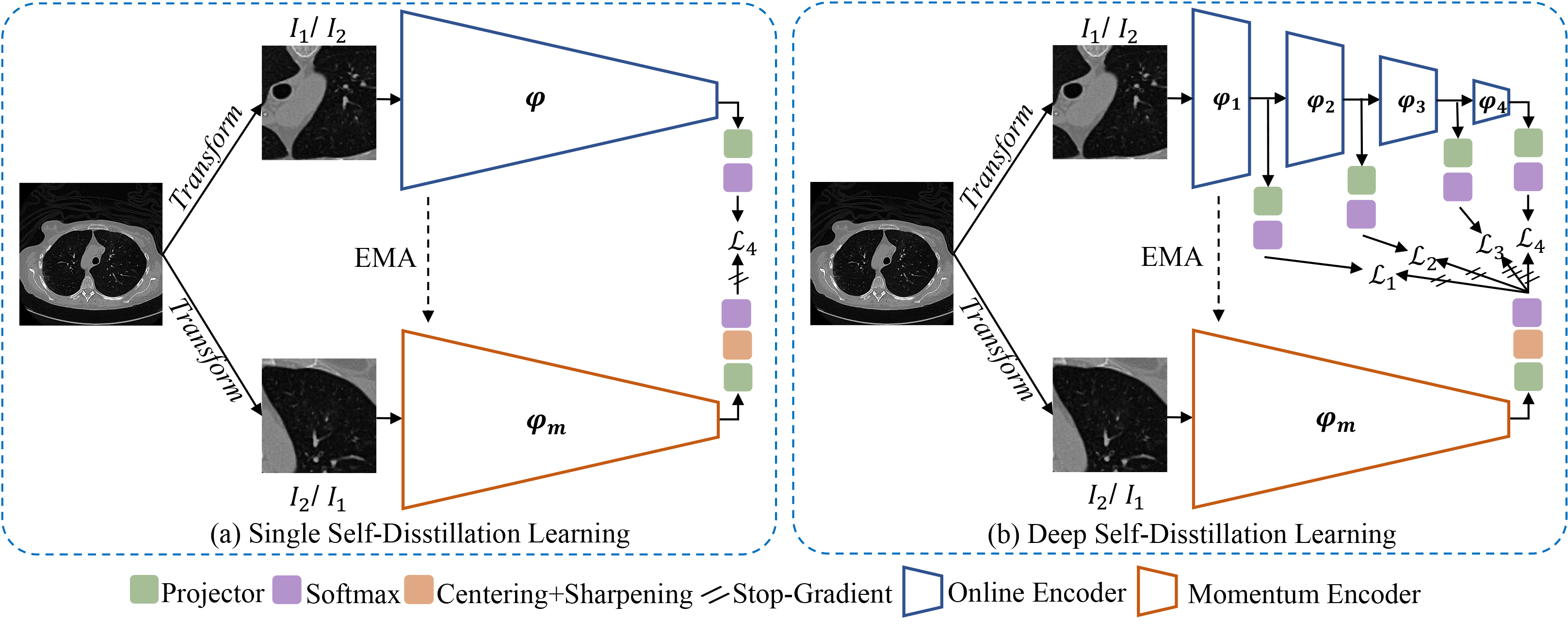
This is the official pytorch implementation of our MICCAI 2022 paper "DeSD: Self-Supervised Learning with Deep Self-Distillation for 3D Medical Image Segmentation". In this paper, we reformulate SSL in a Deep Self-Distillation (DeSD) manner to improve the representation quality of both shallow and deep layers.
This paper is available here.
CUDA 10.1
Python 3.6
Pytorch 1.7.1
Torchvision 0.8.2
- Clone this repo.
git clone https://github.com/yeerwen/DeSD.git
cd DeSD
- Download DeepLesion dataset.
- Run
DL_save_nifti.py(from downloaded files) to transfer the PNG image to the nii.gz form. - Run
re_spacing_ITK.pyto resample CT volumes. - Run
splitting_to_patches.pyto extract about 125k sub-volumes, and the pre-processed dataset will be saved inDL_patches_v2/.
- Run
sh run_ssl.shfor self-supervised pre-training.
- Pre-trained model is available in DeSD_Res50.
As for the target segmentation tasks, the 3D model can be initialized with the pre-trained encoder using the following example:
import torch
from torch import nn
# build a 3D segmentation model based on resnet50
class ResNet50_Decoder(nn.Module):
def __init__(self, Resnet50_encoder, skip_connection, n_class=1, pre_training=True, load_path=None):
super(ResNet50_Decoder, self).__init__()
self.encoder = Resnet50_encoder
self.decoder = Decoder(skip_connection)
self.seg_head = nn.Conv3d(n_class, kernel_size=1)
if pre_training:
print('loading from checkpoint ssl: {}'.format(load_path))
w_before = self.encoder.state_dict()['conv1.weight'].mean()
pre_dict = torch.load(load_path, map_location='cpu')['teacher']
pre_dict = {k.replace("module.backbone.", ""): v for k, v in pre_dict.items()}
# print(pre_dict)
model_dict = self.encoder.state_dict()
pre_dict_update = {k:v for k, v in pre_dict.items() if k in model_dict}
print("[pre_%d/mod_%d]: %d shared layers" % (len(pre_dict), len(model_dict), len(pre_dict_update)))
model_dict.update(pre_dict_update)
self.encoder.load_state_dict(model_dict)
w_after = self.encoder.state_dict()['conv1.weight'].mean()
print("one-layer before/after: [%.8f, %.8f]" % (w_before, w_after))
else:
print("TFS!")
def forward(self, input):
outs = self.encoder(input)
decoder_out = self.deocder(outs)
out = self.seg_head(decoder_out)
return outIf this code is helpful for your study, please cite:
@article{DeSD,
title={DeSD: Self-Supervised Learning with Deep Self-Distillation for 3D Medical Image Segmentation},
author={Yiwen Ye, Jianpeng Zhang, Ziyang Chen, and Yong Xia},
booktitle={Medical Image Computing and Computer Assisted Intervention -- MICCAI 2022},
pages={545--555},
year={2022}
}
Part of codes is reused from the DINO. Thanks to Caron et al. for the codes of DINO.
Yiwen Ye ([email protected])