We propose a novel method for addressing blind source separation of non-linear mixtures via multi-encoder single-decoder autoencoders with fully self-supervised learning. During training, our method unmixes the input into the multiple encoder output spaces and then remixes these representations within the single decoder for a simple reconstruction of the input. Then to perform source inference we introduce a novel encoding masking technique whereby masking out all but one of the encodings enables the decoder to estimate a source signal. To achieve consistent source separation, we also introduce a so-called sparse mixing loss for the decoder that encourages sparsity between the unmixed encoding spaces throughout and a so-called zero reconstruction loss on the decoder that assists with coherent source estimations. We conduct experiments on a toy dataset, the triangles & circles dataset, and with real-world biosignal recordings from a polysomnography sleep study for extracting respiration.
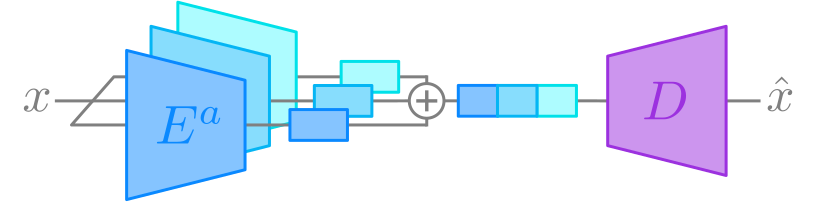
Figure 1a. Training procedure.
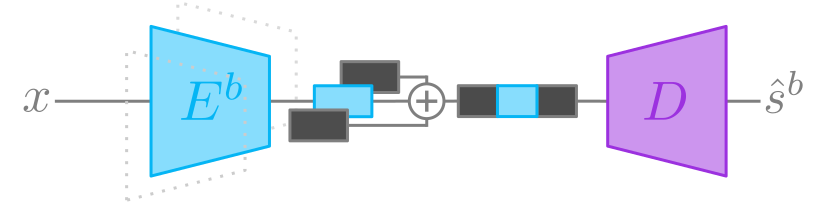
Figure 1b. Inference procedure (source estimation).
As the foundation of our proposed method, we use multi-encoder autoencoders such that each encoder receives the same input, and the outputs of each encoder are concatenated along the channel dimension before being propagated through the single decoder network. During the training phase, a reconstruction loss between the input and output is applied in the typical autoencoder fashion. In addition, we propose two novel regularization methods and a novel encoding masking technique for inference. These three contributions are outlined below...
To estimate a source (i.e. separate a source) with a trained model the
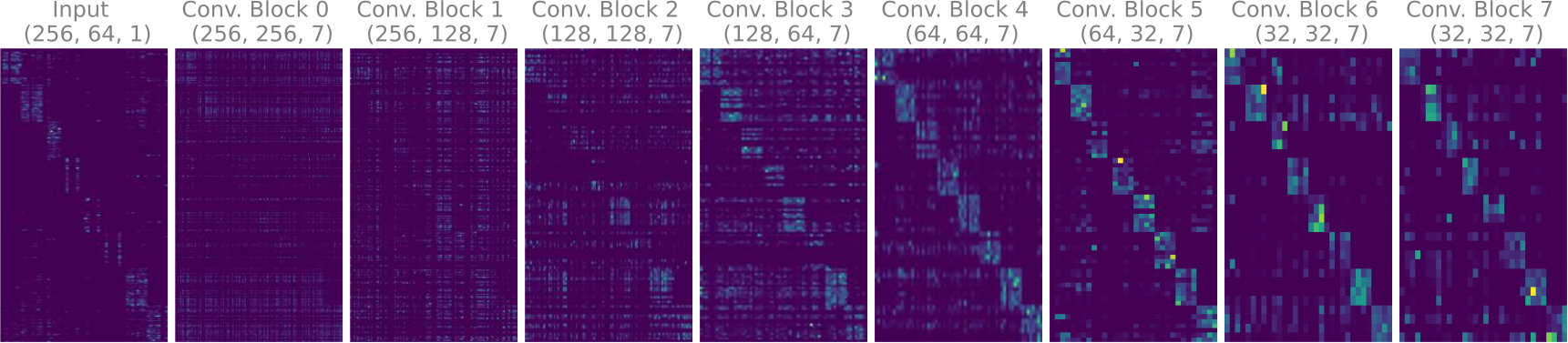
The sparse mixing loss is applied with respect to the channel dimension for each layer's weight
Figure 2. The final decoder weights of the multi-encoder autoencoder model trained on the ECG data are visualized above. The absolute values of the weights are summed along the spatial dimensions to show the effect of the sparse mixing loss.
The zero reconstruction loss is proposed to ensure that masked source encodings have minimal contribution to the final source estimation. For the zero reconstruction loss, an all-zero encoding vector
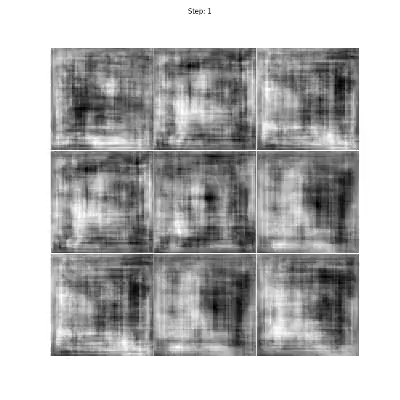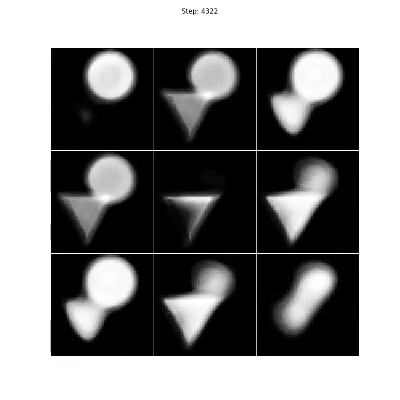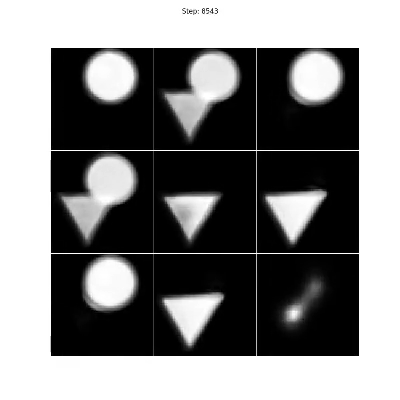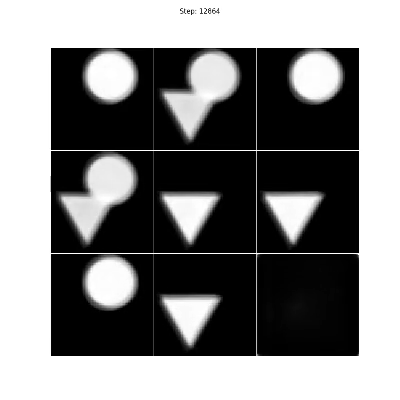
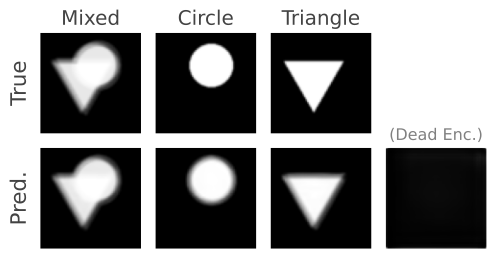
The triangles & circles dataset consists of non-linear mixtures of triangle and circle shapes with uniform random position in size and position. To generate your own dataset please see: notebooks/triangles_and_circles_dataset.ipynb
- To train a model with our configuration use the following command:
python trainer.py experiment_config=tri_and_circ_bss - To test your model please see: notebooks/triangles_and_circles_model_test.ipynb
- Please note that the last checkpoint may not be the best model version. Though we have tried to address the issue of stability (as discussed within the text), instabilities during training may still lead to undesirable results at any point during training. We recommend evaluating as many of the checkpoints as possible to find the best one. Within the output folder of a specific run, there is a
plotsfolder that contains source prediction plots created at set intervals during training to help aid with this process.
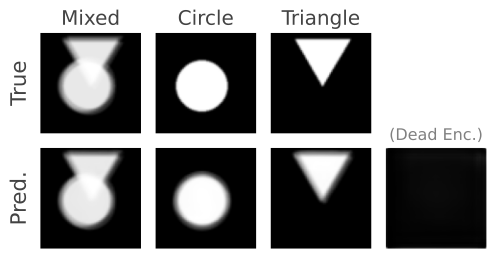
Figure 3.
Figure 4. Even though there are two sources in the mixtures, we choose three encoders to show that the number of sources can be overestimated as the proposed method will converge on a solution where only two of the encoders are responsible for separating the triangles and circles.
You can request access to the Multi-Ethnic Study of Atherosclerosis (MESA) Sleep study12 data here. After downloading the dataset, use the PyEDFlib library to extract the ECG, PPG, thoracic excursion, and nasal pressure signals from each recording. We then randomly choose 1,000 recordings for our training(and validation) and testing splits (45%, 5%, and 50% respectively). Then for each data split we extract segments (each segment with the four simultaneously measured biosignals of interest) with length 12288 as NumPy arrays, resampling each signal to 200hz. At this point, you may use a library for removing bad samples such as the NeuroKit2 library3. We then pickle a list of our segments for ECG and for PPG for both training and testing splits. This file can then be passed to our dataloader (see utils/dataloader/mesa.py) via a setting in the config files. We do not provide this processing code as it is specific to our NAS and compute configuration.
After the data processing is complete and the configuration files are updated with the proper data path (see config/experiment_config/), you can train a model for the ECG or PPG experiments with the following commands:
python trainer.py experiment_config=mesa_ecg_bss
python trainer.py experiment_config=mesa_ppg_bss
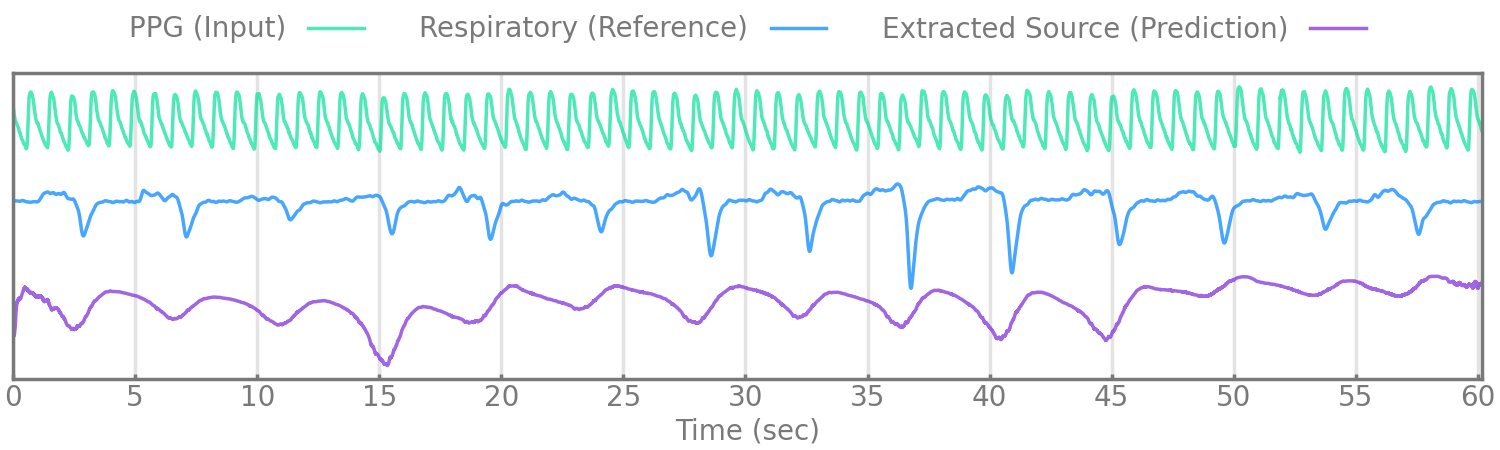
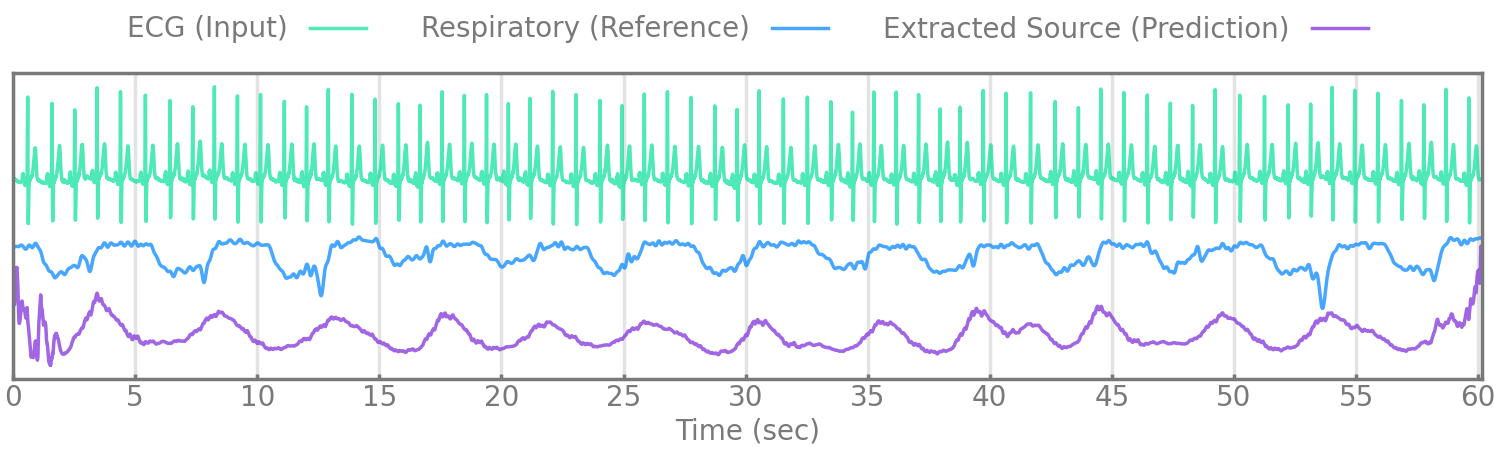
Figure 5.
We evaluate our method by extracting respiratory rate from the estimated source (given by the encoder manually reviewed to correspond with respiration) and comparing it to the extracted respiratory rate of a simultaneously measured reference respiratory signal, nasal pressure, or thoracic excursion.
| Method (Input) | Breaths/Min. MAE |
Breaths/Min. MAE |
Method (Input) | Breaths/Min. MAE |
Breaths/Min. MAE |
|---|---|---|---|---|---|
| BSS | Nasal Pressure | Thoracic Excursion | Heuristic | Nasal Pressure | Thoracic Excursion |
| Ours (PPG) | 1.51 | 1.50 | Muniyandi & Soni, 20174 (ECG) | 2.38 | 2.18 |
| Ours (ECG) | 1.73 | 1.59 | Charlton et al., 20165 (ECG) | 2.38 | 2.17 |
| van Gent et al., 20196 (ECG) | 2.27 | 2.05 | |||
| Sarkar, 20157 (ECG) | 2.26 | 2.07 |
| Method (Input) | Breaths/Min. MAE |
Breaths/Min. MAE |
Method (Input) | Breaths/Min. MAE |
Breaths/Min. MAE |
|---|---|---|---|---|---|
| Supervised (Nasal Presssure as Target) | Nasal Pressure | Thoracic Excursion | Direct Comparison | Nasal Pressure | Thoracic Excursion |
| AE (PPG) | 0.46 | 2.07 | Thoracic Excursion | 1.33 | -- |
| AE (ECG) | 0.48 | 2.16 |
If you find this repository helpful, please cite us.
@misc{webster2024blind,
title={Blind Source Separation of Single-Channel Mixtures via Multi-Encoder Autoencoders},
author={Matthew B. Webster and Joonnyong Lee},
year={2024},
eprint={2309.07138},
archivePrefix={arXiv},
primaryClass={eess.SP}
}
This work was supported by the Technology Development Program (S3201499) funded by the Ministry of SMEs and Startups (MSS, Korea).
Footnotes
-
Zhang GQ, Cui L, Mueller R, Tao S, Kim M, Rueschman M, Mariani S, Mobley D, Redline S. The National Sleep Research Resource: towards a sleep data commons. J Am Med Inform Assoc. 2018 Oct 1;25(10):1351-1358. doi: 10.1093/jamia/ocy064. PMID: 29860441; PMCID: PMC6188513. ↩
-
Chen X, Wang R, Zee P, Lutsey PL, Javaheri S, Alcántara C, Jackson CL, Williams MA, Redline S. Racial/Ethnic Differences in Sleep Disturbances: The Multi-Ethnic Study of Atherosclerosis (MESA). Sleep. 2015 Jun 1;38(6):877-88. doi: 10.5665/sleep.4732. PMID: 25409106; PMCID: PMC4434554. ↩
-
Makowski, D., Pham, T., Lau, Z.J. et al. NeuroKit2: A Python toolbox for neurophysiological signal processing. Behav Res 53, 1689–1696 (2021). https://doi.org/10.3758/s13428-020-01516-y ↩
-
M. Muniyandi, R. Soni, Breath rate variability (brv) - a novel measure to study the meditation effects, International Journal of Yoga Accepted (01 2017). doi:10.4103/ijoy.IJOY_27_17. ↩
-
P. H. Charlton, T. Bonnici, L. Tarassenko, D. A. Clifton, R. Beale, P. J. Watkinson, An assessment of algorithms to estimate respiratory rate from the electrocardiogram and photoplethysmogram Physiological Measurement 37 (4), (2016) 610. doi:10.1088/0967-3334/37/4/610. https://dx.doi.org/10.1088/0967-3334/37/4/610 ↩
-
P. van Gent, H. Farah, N. van Nes, B. van Arem, Heartpy: A novel heart rate algorithm for the analysis of noisy signals, Transportation Research Part F: Traffic Psychology and Behaviour 66 (2019) 368–378. doi: https://doi.org/10.1016/j.trf.2019.09.015. https://www.sciencedirect.com/science/article/pii/S1369847818306740 ↩
-
S. Sarkar, Extraction of respiration signal from ecg for respiratory rate estimation, IET Conference Proceedings (2015) 58 (5 .)–58 (5 .)(1). https://digital-library.theiet.org/content/conferences/10.1049/cp.2015.1654 ↩