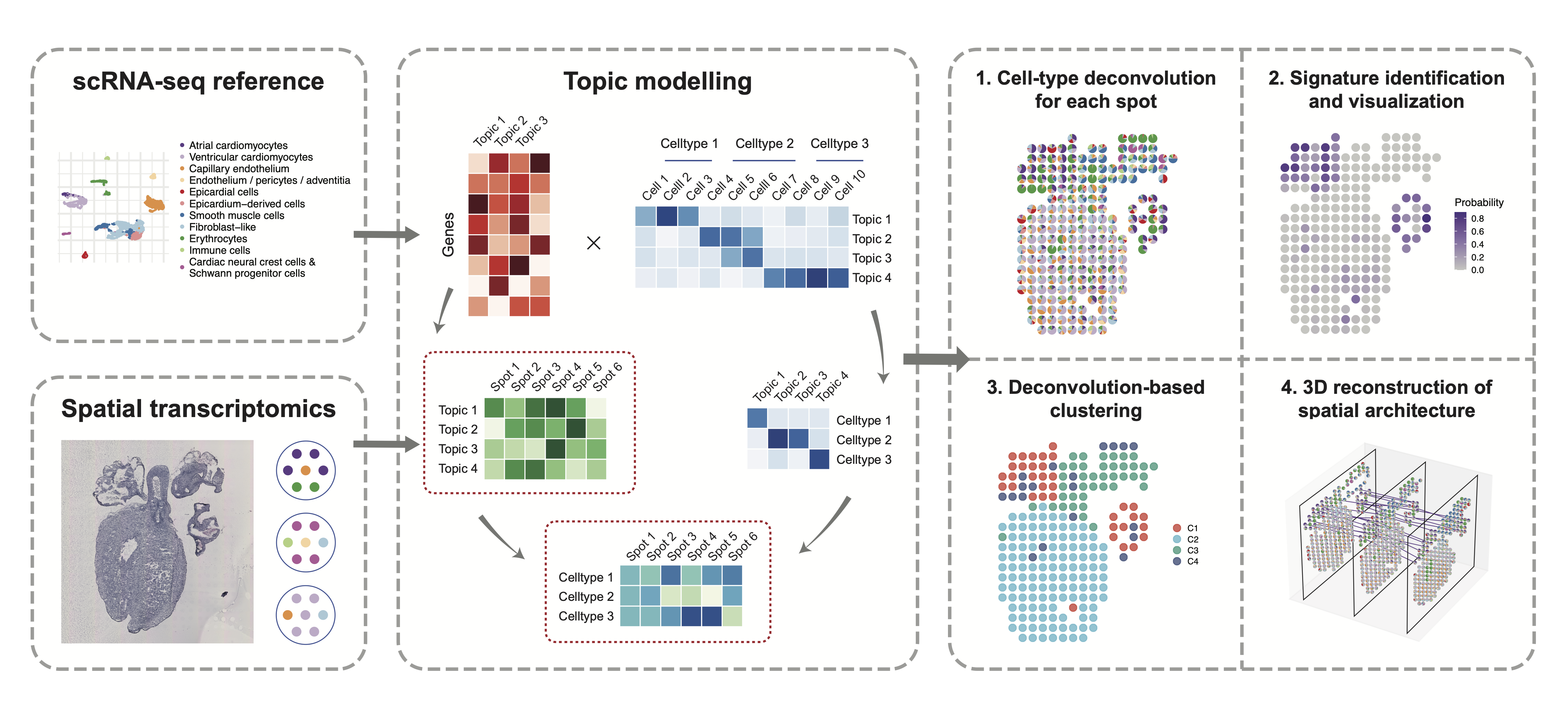
Spatial TRanscrIptomics DEconvolution by topic modeling (STRIDE), is a computational method to decompose cell types from spatial mixtures by leveraging topic profiles trained from single-cell transcriptomics. Besides the cell-type composition deconvolution, STRIDE also provides several downstream analysis functions, including (1) signature (i.e., topic) detection and visualization, (2) spatial clustering and domain identification based on neighborhood cell populations and (3) reconstruction of three-dimensional architecture from sequential ST slides of the same tissue.
For full installation and usage of STRIDE, please refer to the documentation.
- Build STRIDE.
- Add mapping function to identify similarest cells for spatial spots.
- Fix bugs of integration.
- Fix bugs of deconvolution with trained topic model.
- Update mapping function to make cell mapping consistent with the deconvolved fractions.
git clone https://github.com/DongqingSun96/STRIDE.git
cd STRIDE
pip install -r requirements.txt
python setup.py installSTRIDE --help
usage: STRIDE [-h] [-v] {deconvolve,plot,cluster,integrate,map} ...
STRIDE (Spatial TRanscrIptomics DEconvolution by topic modelling) is a cell-
type deconvolution tool for spatial transcriptomics by using single-cell
transcriptomics data.
positional arguments:
{deconvolve,plot,cluster,integrate}
deconvolve Decompose celltype proportion for spatial
transcriptomics.
plot Visualize the deconvolution result.
cluster Neighbourhood analysis based on cell-type composition
and local cell population
integrate Integrate multiple samples from the same tissue.
map Identify similarest cells for spatial spots.
optional arguments:
-h, --help show this help message and exit
-v, --version Print version info.