R tutorial for cleaning data
This tutorial provides some strategies for handling issues with data that need to be resolved before they can be effectively used in visualization and analysis.
Shared RStudioCloud version that you can copy into your account to work with: https://rstudio.cloud/project/3081275
Deal with issues that may come up when importing data files
- Identify and correct structural issues in the source data that prevent clean import into R data structures
- Check and handle data type errors
- Check and handle missing data values
Tuning up the structure of the data to facilitate analysis
- Split up fields that contain mutiple values in a single field
- Check for anomalous values and otherwise explore the data to become familiar with its content and structure.
The data used in this workshop are a modified version of the idigbio_rodents.csv dataset. The data have been modified to illustrate some common problems with datasets that will be addressed in the workshop.
Prior to the workshop you will need to perform the following steps:
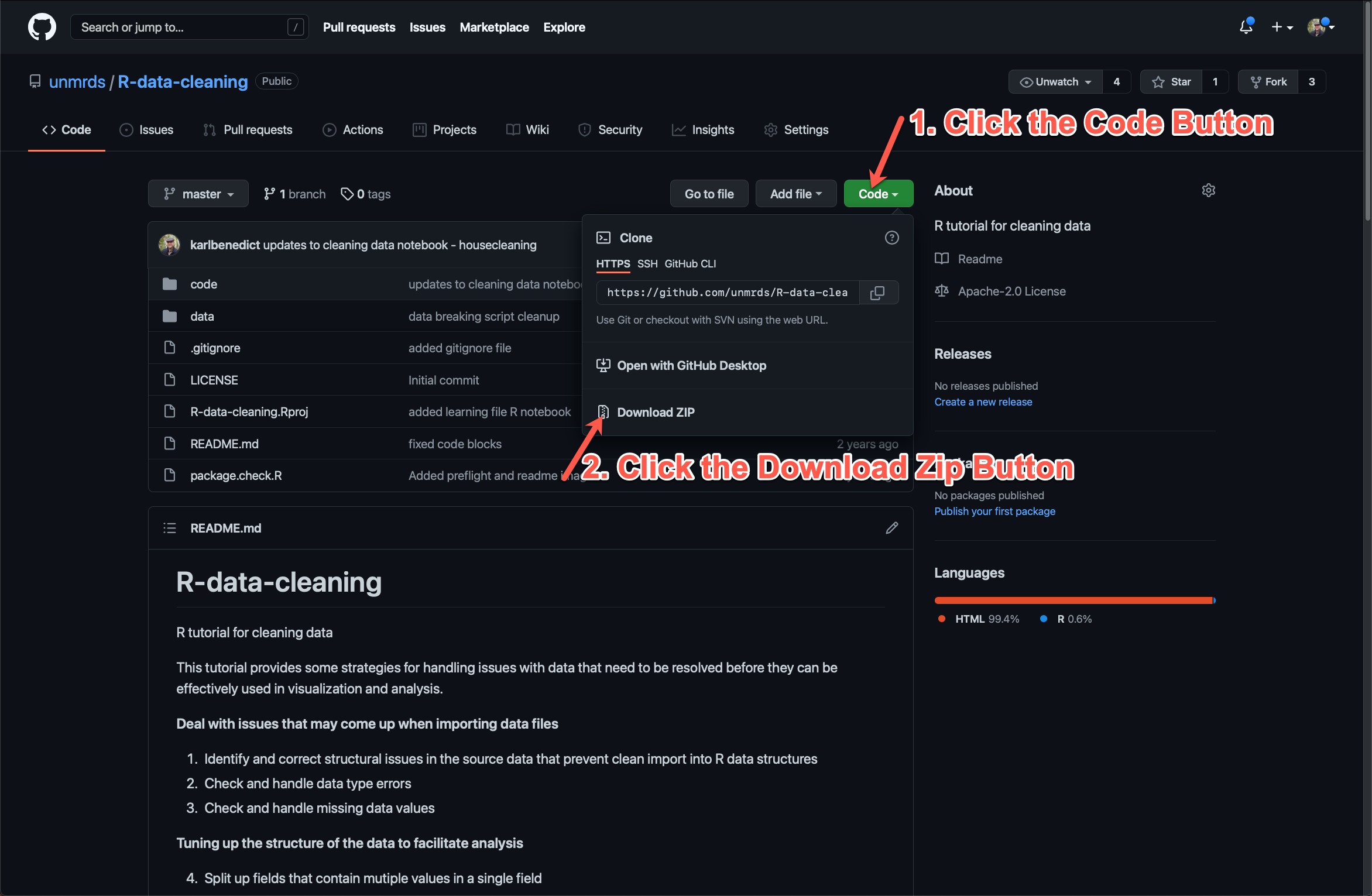
- Download this repository to a folder on your local computer, either as a ZIP archive (see figure below illustrating where to click in the GitHub repository to download the zipfile) or through
git cloneon your computer. If you download the repository as a ZIP archive you will need to uncompress it to work with the collection of files.
-
Install the most recent version of R supported by your operating system from the Comprehensive R Archive Network (CRAN). If you already have R installed on your computer you might consider updating it to a more recent version if it is very old.
-
Install the most recent version of RStudio Desktop for your operating system.
-
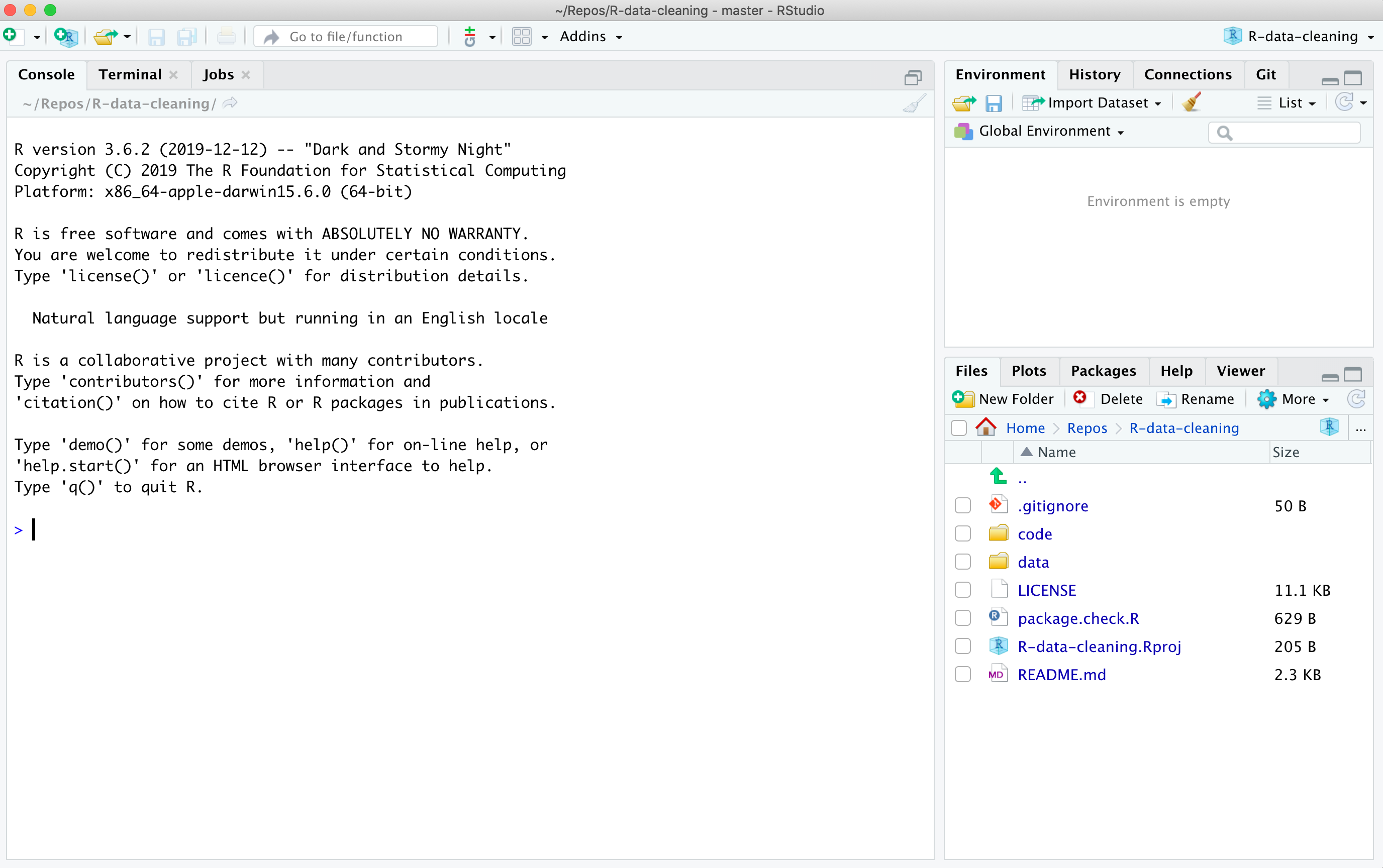
Test your installation of RStudio by opening it through the "Start Menu" in Windows, or the "Applications" folder on a Mac. If you installation was successful in linking R project files with RStudio you can also double-click the
R-data-cleaning.Rprojfile in the top folder of the uncompressed ZIP archive that you downloaded in step (1) above. If you have succeessfully installed and run RStudio you should see a window that looks like the following:
- Install the required R packages (if they are not already installed on your system - if they are already installed you will be told this when executing these commands) by executing the following commands in the R Console:
install.packages("tidyverse")
install.packages("mice")
install.packages("VIM")
install.packages("assertr")- Test that the packages have been properly installed on your system and can be loaded by running the
package.check.Rscript in the top folder of the workshop R project. You can run the script by executing the following command from the R Console:
source('package.check.R', echo=TRUE)If you have successfully installed the required libraries you will see output that looks similar to the following:
> #code by vikram b baliga
> #https://www.vikram-baliga.com/blog/2015/7/19/a-hassle-free-way-to-verify-that-r-packages-are-installed-and-loaded
>
> # .... [TRUNCATED]
> #use this function to check if each package is on the local machine
> #if a package is installed, it will be loaded
> #if any are not, the missing p .... [TRUNCATED]
> #verify they are loaded
> search()
[1] ".GlobalEnv" "package:assertr" "package:VIM" "package:data.table" "package:grid"
[6] "package:colorspace" "package:mice" "package:forcats" "package:stringr" "package:dplyr"
[11] "package:purrr" "package:readr" "package:tidyr" "package:tibble" "package:ggplot2"
[16] "package:tidyverse" "tools:rstudio" "package:stats" "package:graphics" "package:grDevices"
[21] "package:utils" "package:datasets" "package:methods" "Autoloads" "package:base" You should see the following entries in the output of the final search() command in the outpout:
"package:assertr"
"package:VIM"
"package:mice"
"package:tidyverse"If you hadn't previously installed the needed packages you might also see output related to installation and loading of the packages.
The workshop materials are found in two directories within the downloaded repository. Specifically, the data that we will be working with are found in the data folder, and the sample code we will be working through is found in the code folder. The specific files that we will be working with are:
data/learning_struct.csv # for working through structural problems in sourc data files
data/learning.csv # for the rest of the practice, representing source data for which the structural issues have been resolved
code/cleaning_data.Rmd # the R markdown version of the workshop content from which other representations can be generated
code/cleaning_data.html # an HTML rendering of the R markdown sample code and output that is more readable
doce/cleaning_data.R # an R script containing only the executed R code extracted from the R markdown `cleaning_data.Rmd`
The additional files in the data and code directories consist of the source CSV file (data/rawData.csv) from which the data file with problems was generated, and the R code (code/breaking_data.Rmd and the associated code/breaking_data.html) that was used to generate the problem files used in the workshop.