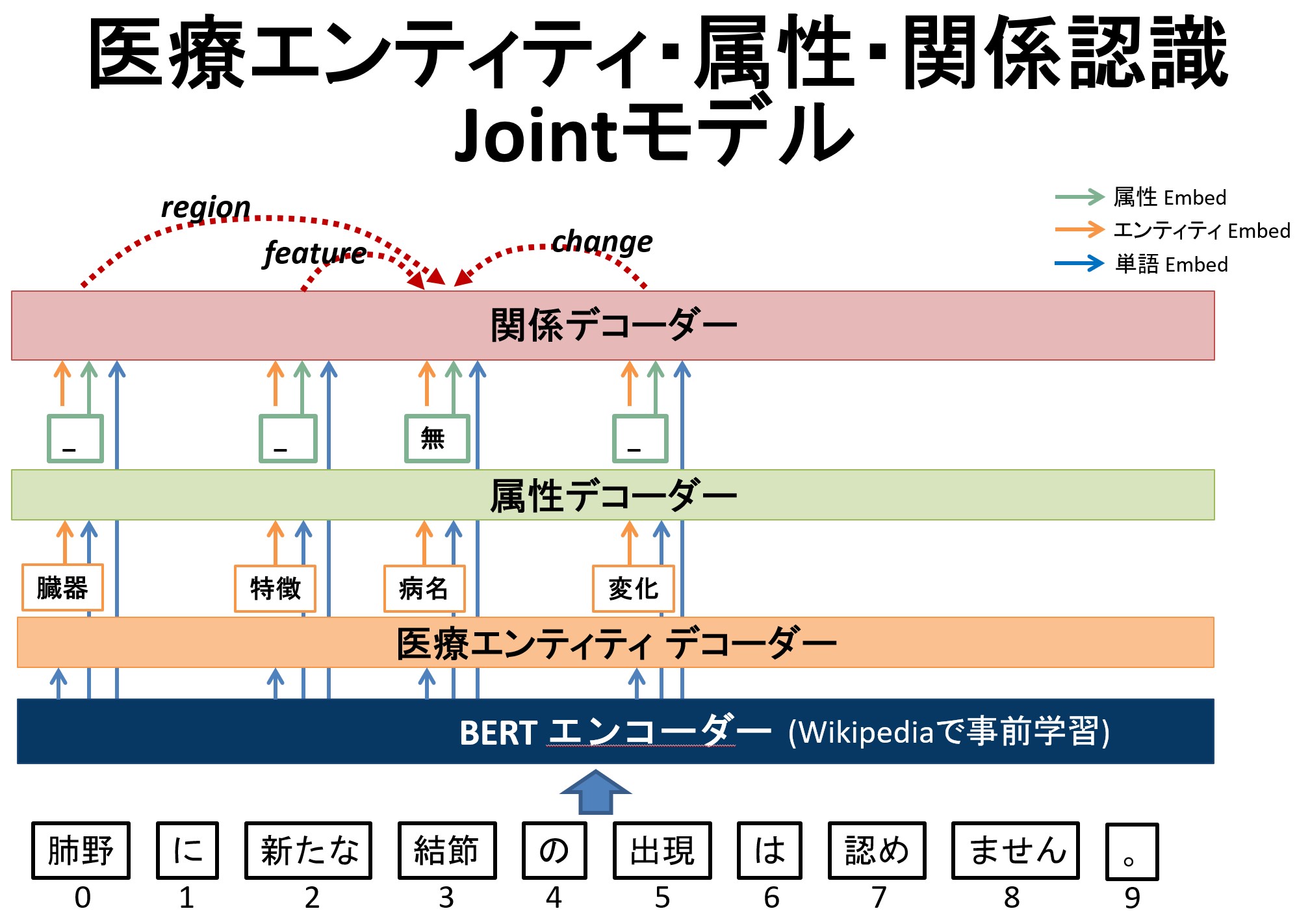
In the field of Japanese medical information extraction, few analyzing tools are available and relation extraction is still an under-explored topic. In this paper, we first propose a novel relation annotation schema for investigating the medical and temporal relations between medical entities in Japanese medical reports. We design a system with three components for jointly recognizing medical entities, classifying entity modalities, and extracting relations.
git clone https://github.com/racerandom/JaMIE.git
cd JaMIE \
pip install -r requirements.txt
mecab (juman-dict) by default
jumanpp
NICT-BERT (NICT_BERT-base_JapaneseWikipedia_32K_BPE)
The Train/Test phrases require all train, dev, test file converted to CONLL-style before Train/Test. You also need to convert raw text to CONLL-style for prediction, but please make sure the file extension is .xml.
python data_converter.py \
--mode xml2conll \
--xml $XML_FILES_DIR \
--conll $OUTPUT_CONLL_DIR \
--cv_num 0 \ # 0 presents to generate single conll file, 5 presents 5-fold cross-validation
--doc_level \ # generate document-level ([SEP] denotes sentence boundaries) or sentence-level conll files
--segmenter mecab \ # please use mecab and NICT bert currently
--bert_dir $PRETRAINED_BERT # Pre-trained BERT or Trained model
CUDA_VISIBLE_DEVICES=$GPU_ID python clinical_joint.py \
--pretrained_model $PRETRAINED_BERT \ # downloaded pre-trained NICT BERT
--train_file $TRAIN_FILE \
--dev_file $DEV_FILE \
--dev_output $DEV_OUT \
--saved_model $MODEL_DIR_TO_SAVE \ # the place to save the model
--enc_lr 2e-5 \
--batch_size 4 \ # depends on your GPU memory
--warmup_epoch 2 \
--num_epoch 20 \
--do_train \
--fp16 (apex required)
We share the models trained on radiography interpretation reports of Lung Cancer (LC) and general medical reports of Idiopathic Pulmonary Fibrosis (IPF):
- The trained model of radiography interpretation reports of Lung Cancer (肺がん読影所見)
- The trained model of case reports of Idiopathic Pulmonary Fibrosis (IPF診療録)
You can either train a new model on your own training data or use our shared model for test.
CUDA_VISIBLE_DEVICES=$GPU_ID python clinical_joint.py \
--saved_model $SAVED_MODEL \ # Where the trained model placed
--test_file $TEST_FILE \
--test_output $TEST_OUT \
--batch_size 4
python data_converter.py \
--mode conll2xml \
--xml $XML_OUT_DIR \
--conll $TEST_OUT
We offer the links of both English and Japanese annotation guidelines.
Recognition accuracy can be improved by leverage more training data or more robust pre-trained models. We are working on making the code compatible with Japanese DeBERTa
If you have any questions related to the code or papers, please feel free to send a mail to Fei Cheng: [email protected] or [email protected]
If you use our code in your research, please cite the following papers:
@inproceedings{cheng-etal-2022-jamie,
title={JaMIE: A Pipeline Japanese Medical Information Extraction System with Novel Relation Annotation},
author={Fei Cheng, Shuntaro Yada, Ribeka Tanaka, Eiji Aramaki, Sadao Kurohashi},
booktitle={Proceedings of the Thirteenth Language Resources and Evaluation Conference (LREC 2022)},
year={2022}
}
@inproceedings{cheng2021jamie,
title={JaMIE: A Pipeline Japanese Medical Information Extraction System},
author={Fei Cheng, Shuntaro Yada, Ribeka Tanaka, Eiji Aramaki, Sadao Kurohashi},
booktitle={arXiv},
year={2021}
}
@inproceedings{yada-etal-2020-towards,
title={Towards a Versatile Medical-Annotation Guideline Feasible Without Heavy Medical Knowledge: Starting From Critical Lung Diseases},
author={Shuntaro Yada, Ayami Joh, Ribeka Tanaka, Fei Cheng, Eiji Aramaki, Sadao Kurohashi},
booktitle={Proceedings of the Twelfth Language Resources and Evaluation Conference (LREC 2020)},
year={2020}
}
@inproceedings{cheng-etal-2020-dynamically,
title={Dynamically Updating Event Representations for Temporal Relation Classification with Multi-category Learning},
author={Fei Cheng, Masayuki Asahara, Ichiro Kobayashi, Sadao Kurohashi},
booktitle={Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP 2020), Findings Volume},
year={2020}
}