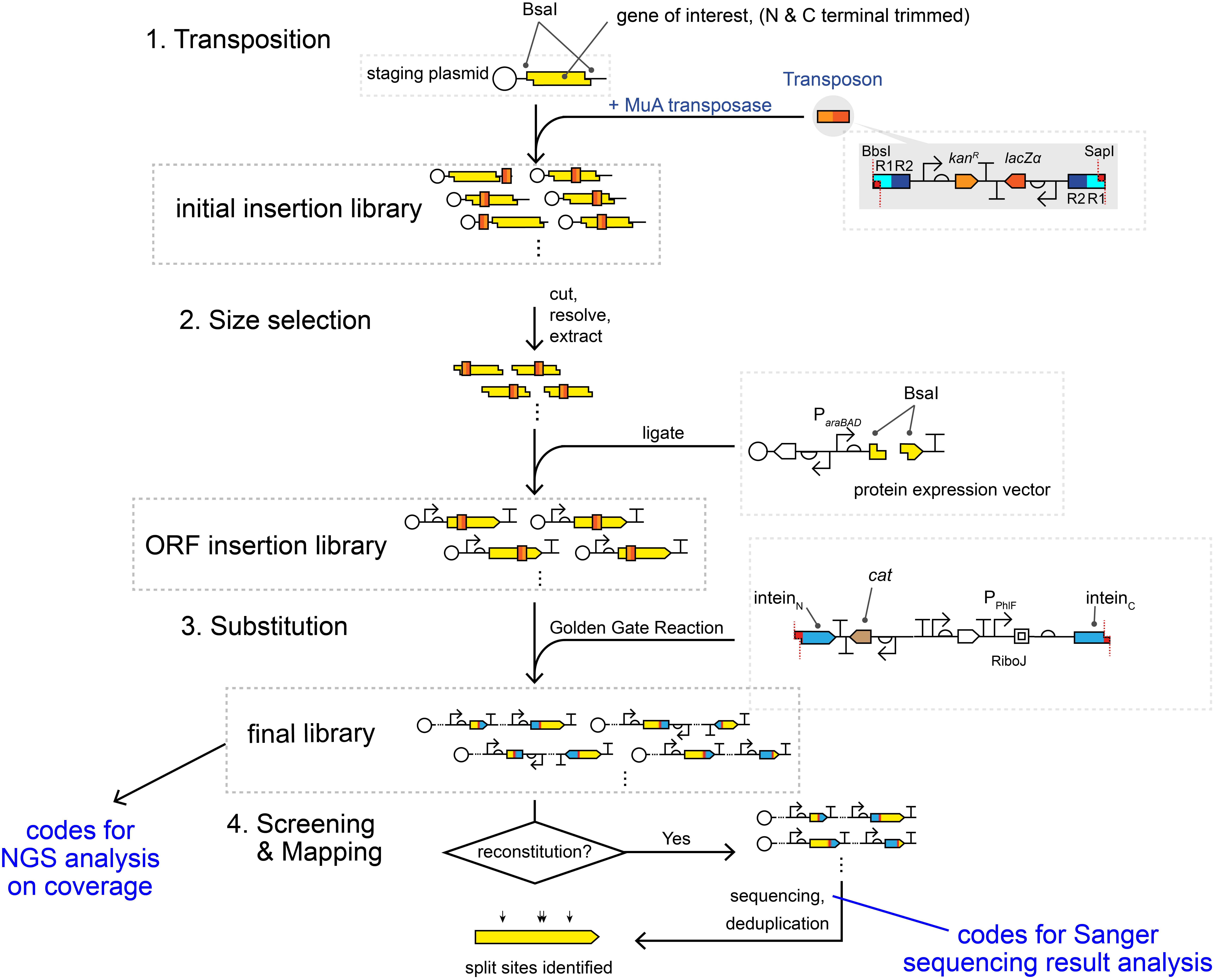
Manuscript: "Intein-assisted bisection mapping systematically splits proteins for Boolean logic and inducibility engineering"
Repository for codes used in IBM experiments.
For analyzing Sanger sequencing results at the last stage of PCR products, please refer to the notebook:
identify_split_site.ipynb
For analyzing NGS data on final IBM libraries, please refer to the manual:
NGS_analysis manual.ipynb
which will provide an overview of steps
Note: For NGS analysis, check out dipseq for a better pipeline.
- Pandas
- Biopython
Licensed under the MIT License.