-
The overall aim of the second part of the project is to improve the U-Net segmentation by incorporating synthetic data (generated with a generative model) in the U-Net training. Below is a very generic research question to this effect, it is good if you can come up with a more specific research question that you answer in your project.
-
If you complete the VAE code, you will be able to perform unconditional image generation. To help with training a segmentation model you will need to consider how to also get labels for these images (e.g. conditional generation with SPADE layers or using a previous version of your segmentation U-Net, etc.). Additionally, the images generated by the VAE can be lower quality and you can work on ways to improve this.
-
apply_segmentation.py shows an example script to apply a trained segmentation model to one (arbitrarily selected) slice – you would need to extend this to apply it to all slices for a patient.
-
It is advised to train models in 2D. To apply the segmentation model to a new patient in 3D, you can apply it slice by slice in a for loop and store the output again as a 3D image.
-
To save computation cost, you can work with the pre-processed images as I do in the data loader provided (first center cropped to 256x256 and then downsampled to 64x64). For your final results, you can choose to either evaluate at this resolution or to resample and un-crop to the original image space.
-
The slides: 3 - generative models are just for your reference and were not covered during the course. But maybe they might inspire you for your project.
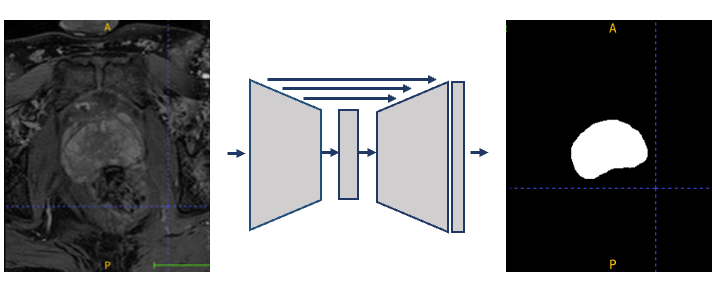
An alternative approach for automatic segmentation, which does not require registration, is via pixel-based classification. Here the delineated patient images form the training set, and each pixel is a sample, labeled as either foreground (organ of interest) or background. The training set is used to train a classifier which predicts for each pixel of the input image whether it is foreground or background. A common approach is to use a deep learning model such as a U-Net, demonstrated in Figure 1.
Figure 1: a schematic of a U-Net model, taking the prostate MR image as input and outputting the binary segmentation mask.
However, for training, it is important to have a large training set, and in practice this may not always be available. A large dataset is not available in this case. A recent idea to tackle this problem is to train deep generative models to generate synthetic training data. These synthetic images can then be used along with the real images in order to train the segmentation model.
How does the integration of synthetic generated images into the training dataset impact the performance of deep learning models for automated prostate segmentation in MRI data? (Think of a suitable metric and compare segmentation performance with and without synthetic data).
To get you started with the project, in the practical session, you will train: Week 1. A segmentation model based on the U-Net. Week 2. A VAE model to generate synthetic data. This will serve as a baseline generative model in the "project" part of this course.
You won't implement the models from scratch. The outline of the code is already given (have a look around the files in the folder code). The parts that you will implement are indicated with # TODO.
First, you will implement the segmentation model. To do this, start from train_unet.py. Once you can train the segmentation model, move on to the VAE.
Note that the model architecture is designed to process 2D images, i.e., each slice is segmented individually. To save computation time, the images are downsampled during loading (this is already implemented).
For your project, you are free to build on top of the practical session or to use a different approach. Remember the discussion from the lecture on the limitations of VAEs and potential solutions to this. Since the focus of this part of the course is on generative models, it is recommended to spend time optimising the gernerative model rather than the segmentation model.
This project is more similar to a research project than a course assignment and as such, there is no guarantee that you will get good results from the model. Your project grade is based on your approach, i.e., you will score highly for a good approach that is well motivated in the report.
- First, you need to download the code. You can either download the code as zip by choosing
Download zipfrom the dropdown menu of the greenCodebutton in this repository or you can clone the repository. - Once you have downloaded the code, create a new python environment and install the required packages. Either use conda or via pip with the python virtual environment manager:
- Open a terminal and navigate into the
codedirectory. - Run
python3 -m venv venvto create a new virtual environment. - Activate the environment. The command for this depeonds on your operating system. For Mac,
source venv/bin/activate, for Windowssource venv/Scripts/activate. - You might need to upgrade pip by
pip install --upgrade pip - Install the dependencies from the
requirements.txtfile:pip install -r requirements.txt.
- Open a terminal and navigate into the
- How to have corresponding labels for your generated images (e.g. conditional generation or using the segmentation model).
- How to increase the variation in your generated data.