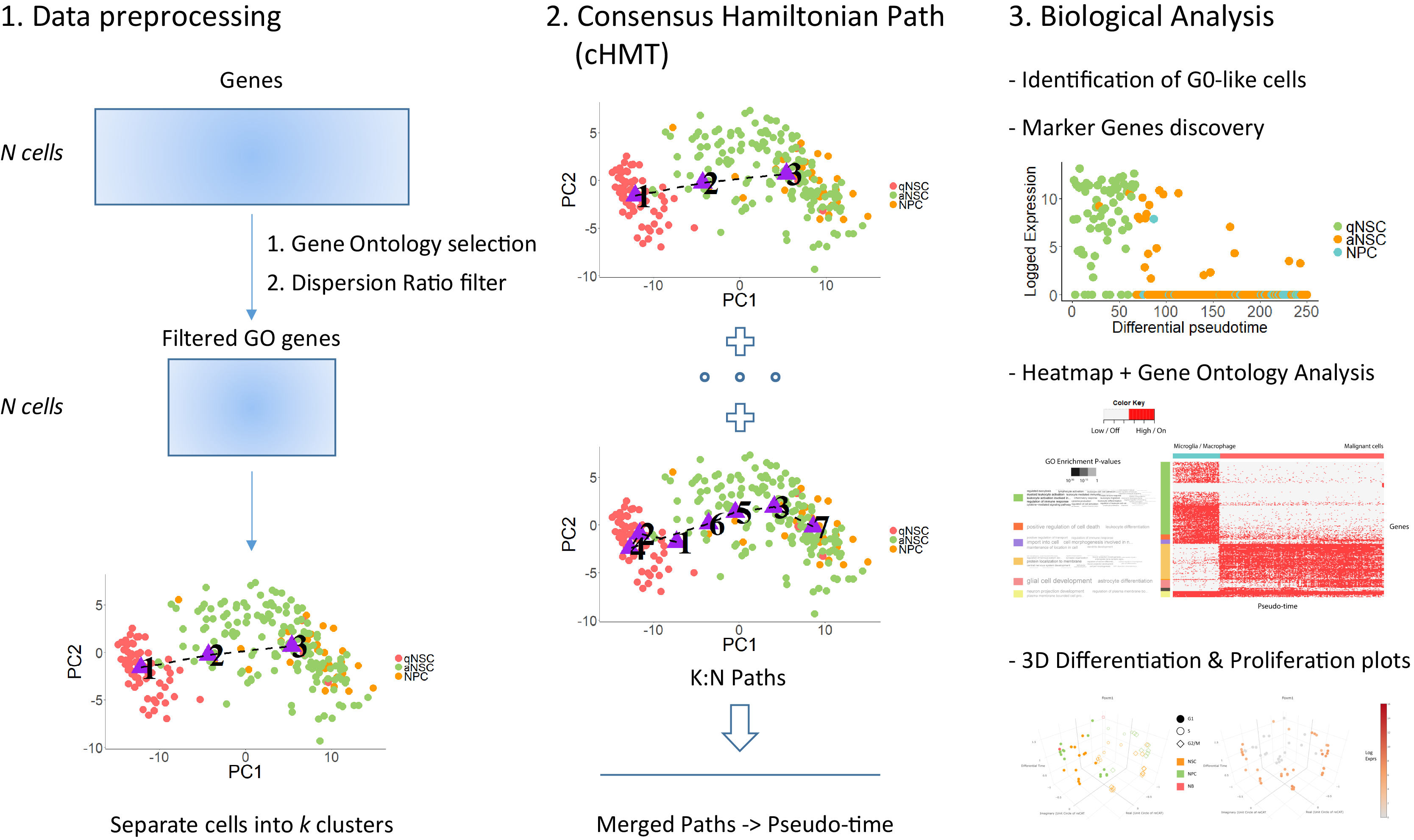
redPATH reconstructs the pseudo development time of cell lineages in single-cell RNA-seq data. It formulates the problem of pseudo temporal ordering into a Hamiltonian path problem and attempts to recover the pseudo development time in single-cell RNA-seq datasets. We provide a comprehensive analysis software tool with robust performance.
car, combinat, doParallel, dplyr, energy, ggplot2, GOsummaries, gplots, MASS, mclust, minerva, plotly, Rcpp, RcppArmadillo, scater
After downloading the package, please extract and rename the folder to "redPATH"
require(devtools)
setwd("where redPATH folder is located")
install("redPATH", args = c("--no-multiarch")
Assuming your input data is a m genes (rows) by n cells (cols) matrix. Here, a neural stem cell (145 cells) dataset from 2015 (1) is used as an example analysis and a diseased example (MGH107 - 252 cells) dataset is also provided from 2017 (2).
library(redPATH)
input_data <- llorens_exprs # A sample single cell dataset
# Reduce genes to ones which are related to Cell Development, Cell Morphology, Cell Differentiation, Cell Fate and Cell Maturation (from Gene Ontology)
subset_GO_data <- get_GO_exp(input_data, gene_type = "gene_names", organism = "mouse", takelog = F)
# Filter genes using a simple dispersion threshold
filtered_GO_data <- filter_exp(subset_GO_data, dispersion_threshold = 10, threads = 2)
An optional function is also available to identify the G0-like cells:
sample_mean_score <- llorens_reCAT_scores
# Get kmeans clusters
kmeans_clusters <- kmeans_clustering(sample_mean_score$mean_scores, k = 5)
test_for_g0 <- statistical_test(sample_mean_score$mean_scores, kmeans_clusters, threshold = 0.001)
# Example output:
# [1] "Possible group of G0-like cells is: 2"
new_input_data <- input_data[,-which(kmeans_clusters == 2)]
# Evaluation plots:
d1 <- distribution_plot(sample_mean_score$mean_scores, kmeans_clusters)
d2 <- mean_score_plot(sample_mean_score$mean_scores, kmeans_clusters, reCAT_ordIndex)
redpath_pseudotime <- redpath(filtered_GO_data, threadnum = 4, base_path_range = c(4:8))
# base_path_range is set to the estimated number of cell types, k0, and k0 +4.
# Plot specific gene expression along the pseudotime
cdk_gene_exprs <- get_specific_gene_exprs(full_gene_exprs_data = input_data, gene_name = "cdk11", type = "match", organism = "mm")
plot_cdk <- plot_specific_gene_exprs(cdk_gene_exprs, redpath_pseudotime, color_label = cell_type_label, order = T)
require(scater)
multiplot(plotlist = plot_cdk, layout = matrix(c(1:2), 2, 1)) # For a total number of 2 plots
# Distance Correlation and Maximal Information Coefficient calculation
dcor_result <- calc_correlation(full_exprs_matrix = input_data, redpath_pseudotime, type = "dcor")
mic_result <- calc_correlation(full_exprs_matrix = input_data, redpath_pseudotime, type = "mic")
# Union Selection
selected_result <- gene_selection(full_exprs_matrix = input_data, redpath_pseudotime, dcor_result, mic_result, threshold = 0.5)
# Intersection Selection
selected_result <- gene_selection_intersection(full_exprs_matrix = input_data, redpath_pseudotime, dcor_result, mic_result, threshold = 0.5)
# selected_result returns a m genes by n cells matrix with the selected significant genes
# Calculating the ON/HIGH or OFF/LOW state of each cell for each gene.
gene_state_result <- calc_all_hmm_diff_parallel(selected_result, cls_num = 2, threadnum = 4)
# Optional: plots the top 10 significant genes
hmm_plots <- plot_hmm_full(selected_result, redpath_pseudotime, gene_state_result, cell_type_label = llorens_labels, num_plots = 10)
require(scater)
multiplot(plotlist = hmm_plots, layout = matrix(c(1:10), 5, 2)) # For a total number of 10 plots
# Infer gene clusters using hierarchical clustering
inferred_gene_cluster <- get_gene_cluster(sorted_matrix = gene_state_result, cls_num = 8)
# Heatmap plot
cell_labels <- llorens_labels
heatmap_plot <- plot_heatmap_analysis(sorted_matrix = inferred_gene_cluster, redpath_pseudotime, cell_labels = llorens_labels, gene_cluster = inferred_gene_cluster, input_type = "hmm")
GO_summary_result <- infer_GO_summaries(inferred_gene_cluster, organism = "mmusculus")
plot(GO_summary_result, fontsize = 8)
# normalizedResultLst can be loaded from the output .RData file from reCAT.
p1 <- plot_cycle_diff_3d(normalizedResultLst, redpath_pseudotime, cell_cycle_labels, cell_type_labels)
p1
# normalizedResultLst can be loaded from the output .RData file from reCAT.
# get specific genes of interest:
cdk_gene_exprs <- get_specific_gene_exprs(input_data, gene_name = "cdk11", type = "match", organism = "mm")
p1 <- plot_diff_3d(normalizedResultLst, redpath_pseudotime, cell_cycle_labels, cdk_gene_exprs)
p1
This work is currently under submission. Reconstructing the pseudo development time of cell lineages in single-cell RNA-seq data and applications in cancer.
References:
1. Llorens-Bobadilla, E., Zhao, S., Baser, A., Saiz-Castro, G., Zwadlo, K. and Martin-Villalba, A. (2015) Single-Cell Transcriptomics Reveals a Population of Dormant Neural Stem Cells that Become Activated upon Brain Injury. Cell Stem Cell, 17, 329-340.
2. Venteicher, A.S., Tirosh, I., Hebert, C., Yizhak, K., Neftel, C., Filbin, M.G., Hovestadt, V., Escalante, L.E., Shaw, M.L., Rodman, C. et al. (2017) Decoupling genetics, lineages, and microenvironment in IDH-mutant gliomas by single-cell RNA-seq. Science, 355.
If there's any questions / problems regarding redPATH, please feel free to contact Ken Xie - [email protected]. Thank you!