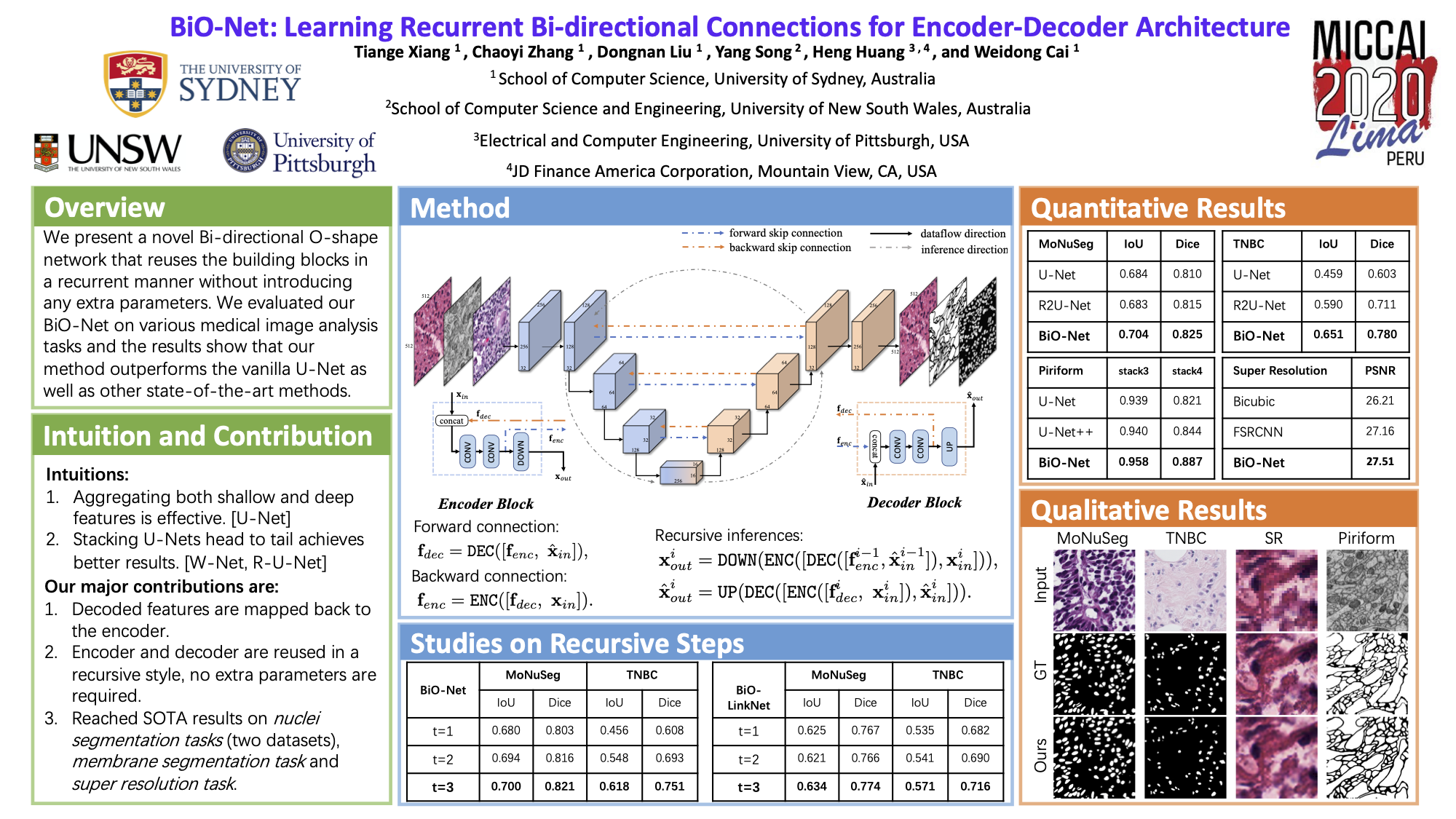
Official implementation of "BiO-Net: Learning Recurrent Bi-directional Connections for Encoder-Decoder Architecture", MICCAI 2020
Paper: https://arxiv.org/abs/2007.00243
News (2021-06-26): Check out our latest work BiX-NAS that is to appear in MICCAI2021! BiX-Net Achieves on par performances to BiO-Net but with 34.9 times fewer parameters and 4.0 times fewer MACs!
- Python >= 3.6
- PIL >= 7.0.0
- matplotlib >= 3.3.1
Keras version
- tensorflow-gpu >= 1.14.0
- keras >= 2.1.5
Pytorch version
- tqdm >= 4.54.1
- imgaug >= 0.4.0
- torch >= 1.5.0
- torchvision >= 0.6.0
NOTE1: Both versions have been sucessfully tested on a GeForce RTX 2080 GPU with CUDA=10.1 and driver=435.21.
NOTE2: You don't need all dependencies to run on a sole backend.
Please refer to the official website (or project repo) for license and terms of usage. We preprocessed the data for adaptating our data loaders. Please find the links below.
MoNuSeg
- Official Website: https://monuseg.grand-challenge.org/Data/
- Baidu Disk: https://pan.baidu.com/s/1tqDzX52v8GYWXF4YfUGu1Q password: dqsr
- Google Drive: https://drive.google.com/file/d/1j7vEoq6YCBNKMoOZKPSQNciHZkVzkxGD/view?usp=sharing
TNBC
- Official Repo: https://github.com/PeterJackNaylor/DRFNS
- Baidu Disk: https://pan.baidu.com/s/1zPWTYAEffX55c2eyb3cU0Q password: zsl1
- Google Drive: https://drive.google.com/file/d/1RYY7vE0LAHSTQXvLx41civNRZvl-2hnJ/view?usp=sharing
Use --backend to switch between backends [keras, pytorch], default=keras.
Train
python3 main.py --epochs 300 --iter 3 --integrate --train_data PATH_TO_TRAIN_DATA_ROOT \
--valid_data PATH_TO_VALID_DATA_ROOT --exp 1 --backend YOUR_BACKEND
Evaluate
Save metrics and segmentations:
python3 main.py --evaluate_only --save_result --valid_data PATH_TO_VALID_DATA_ROOT --exp 1 --backend YOUR_BACKEND
Print metrics only:
python3 main.py --evaluate_only --valid_data PATH_TO_VALID_DATA_ROOT --exp 1 --backend YOUR_BACKEND
NOTE: --iter,--integrate,--multiplier need to be specidied on the above commands.
or
python3 main.py --evaluate_only --valid_data PATH_TO_VALID_DATA_ROOT --model_path PATH_TO_TRAINED_MODEL --backend YOUR_BACKEND
If you find this repo useful in your work or research, please cite:
@inproceedings{xiang2020bio,
title={BiO-Net: Learning Recurrent Bi-directional Connections for Encoder-Decoder Architecture},
author={Xiang, Tiange and Zhang, Chaoyi and Liu, Dongnan and Song, Yang and Huang, Heng and Cai, Weidong},
booktitle={International Conference on Medical Image Computing and Computer-Assisted Intervention},
pages={74--84},
year={2020},
organization={Springer}
}