Preprint manuscript of this method available at https://doi.org/10.1101/2023.04.26.538392
MitoSort is an efficient computational method to demultiplex samples from different individuals and detect cross-genotype doublets using endogenous mtDNA germline variants. It is comprised of 6 steps with the first 2 using external tools and other using in-house script. Users can run each step one by one:
- Realign MT sequences (using GATK, 01_MT_realign.py)
- Find SNP (using Varscan2, 02_find_SNPs.py)
- Divide bam (03_divide_BAM.py)
- Retain cell barcode (04_retain_cell_barcodes.py)
- Generate SNP matrix (05_generate_SNP_matrix.py)
- Demultiplex (06_demultiplex.py)
For the convenience of users, we also encapsulate MitoSort pipeline into three subcommands including mt-realign, generate-snp-matrix and demultiplex. Users can input the name of subcommand for further help information :
Usage: MitoSort_pipeline.py [OPTIONS] COMMAND [ARGS]...
Options:
--help Show this message and exit.
Commands:
demultiplex Identifying cross-genotype doublets and demultiplexing samples
generate-snp-matrix Generate SNP matrices
mt-realign Realign mitochondrial reads using GATKUsers can create a conda environment to install all the required python packages of MitoSort
# clone MitoSort repository
git clone https://github.com/tangzhj/MitoSort.git
# create a conda environment and install all the required python packages
conda env create -f /path/to/MitoSort/MitoSort_env.yaml
conda activate MitoSort
In addition to required python packages. MitoSort also requires GATK for MT realignment and VarScan2 for variant calling. Users should install them and specify the path of tool when running MitoSort. We also upload the versions of tools we have tested in this repository.
First, mitochondrial reads are realigned by GATK by calling mt-realign command. The following are options for mt-realign command. It takes a BAM file as input and output a BAM file containing realigned MT reads (possorted_chrM_realign.bam).
Usage: MitoSort_pipeline.py mt-realign [OPTIONS]
Realign mitochondrial reads using GATK
Options:
-b, --bam_file PATH BAM file output by Cellranger [required]
-f, --genome_fasta PATH FASTA file of reference genome [required]
--gatk_path PATH Path to GenomeAnalysisTK(GATK) [required]
-o, --output_dir PATH Output parent directory [required]
--data_type TEXT Type of data. Default is ATAC. Set to RNA if your
data is scRNA
-h, --help Show this message and exit.
A typical mt-realign command looks like
python MitoSort_pipeline.py mt-realign -b /path/to/possorted_bam.bam -f /path/to/reference.fasta --gatk_path /path/to/GenomeAnalysisTK_3.5-0.jar -o /path/to/output_dir
Given a list of barcodes, SNP matrices containg alternative counts, reference counts and allele frequency are generated for these barcodes by calling generate-snp-matrix command. The following are options for generate-snp-matrix command. Note that the output_dir option should be the same as that of mt-realign command.
Usage: MitoSort_pipeline.py generate-snp-matrix [OPTIONS]
Generate SNP matrices
Options:
-b, --bam_file PATH BAM file containing realigned MT reads
`possorted_chrM_realign.bam` [required]
-f, --genome_fasta PATH FASTA file of reference genome [required]
-c, --cell_barcode PATH A CSV file containing barcode metrics output by
Cellranger
(singlecell.csv/per_barcode_metrics.csv). If it
isn't generated by Cellranger, please input a file
cotaining cell barcodes with header named `barcode`
[required]
-m, --chrm_length PATH a BED file containing chrM region [required]
--varscan_path PATH Path to VarScan [required]
-o, --output_dir PATH Output parent directory [required]
--cell_tag TEXT Default is CR. Set if your cell barcode tag is not
CR
-h, --help Show this message and exit.
A typical generate-snp-matrix command looks like
python MitoSort_pipeline.py generate-snp-matrix -b /path/to/possorted_chrM_realign.bam -f /path/to/reference.fasta -c /path/to/singlecell.csv -m /path/to/MitoSort/data/hg38_chrM.bed --varscan_path /path/to/VarScan.v2.3.7.jar -o /path/to/output_dir
The core of MitoSort is to identify cross-genotype doublets and demultiplex samples from different individuals. It can be achieved by calling demultiplex command. Users can tune the parameters and run it multiple times to achieve best performance on specific datasets. The following are options for demultiplex command. Note that the output_dir option should be the same as that of mt-realign command.
Usage: MitoSort_pipeline.py demultiplex [OPTIONS]
Identifying cross-genotype doublets and demultiplexing samples
Options:
-o, --output_dir PATH Output parent directory [required]
-k, --clusters TEXT number of pooled individuals [required]
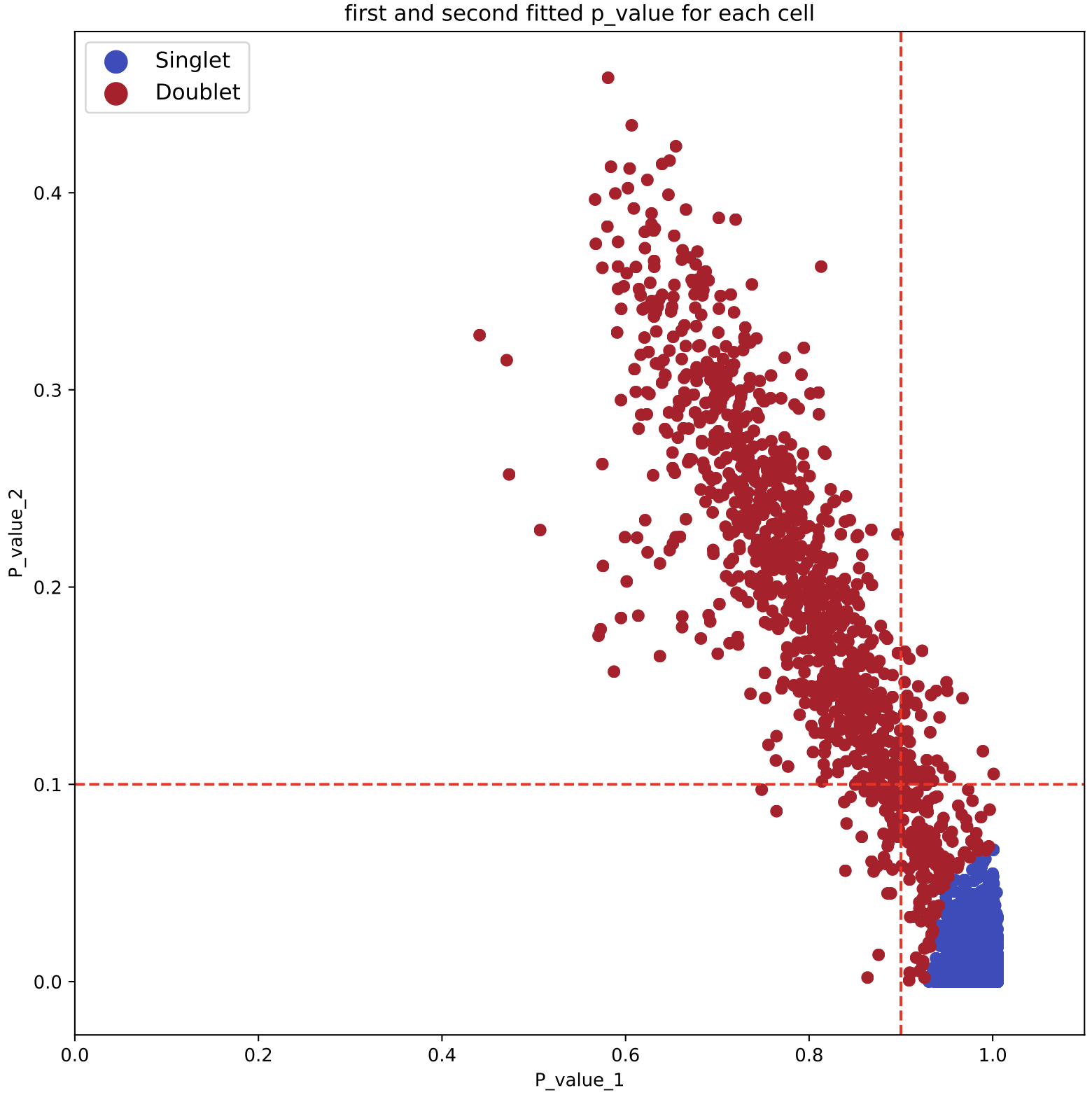
--p1_cutoff TEXT maximum cutoff of p1 for doublet identification.
Default to be 0.9
--p2_cutoff TEXT minimun cutoff of p2 for doublet identification.
Default to be 0.1
--depth_cutoff TEXT If the depth per cell is less than the specified
depth threshold (depth_percell), then assign the value
'unassigned'. Default to be 1.
--method TEXT Default is 'full', if the number of cell varies
greatly in different samples, try 'direct'.
-h, --help Show this message and exit.
A typical demultiplex command looks like
python MitoSort_pipeline.py demultiplex -o /path/to/output_dir -k number_of_pooled_individuals
The important output files are
specific_gemeline.txt(specific mtDNA germline variants for each individual)result_pvalue.txt
The following is example ofresult_pvalue.txt, which includes the cell barcode, singlet/doublet status, assigned sample, p1 and p2:
Barcode Demultiplex P_value_1 P_value_2 Sample0_Pr Sample1_Pr Sample2_Pr Sample3_Pr
LibA_TCACCACTCCCAATAG-1 Sample2 1.0 0.00205835430593 0.00205835430593 0.0 1.0 0.000257875653677
LibA_CGAGTTAGTGATGCTT-1 Sample0 0.998651193644 0.00733731472454 0.998651193644 0.0 0.0 0.00733731472454
LibB_CAGCTGGGTTGCCGCA-1 Sample1 1.0 0.0 0.0 1.0 0.0 0.0
LibC_TAAGTGCGTATTCGCA-1 Doublet 0.590786198879 0.329170270029 0.000912548265621 0.329170270029 0.00440488270379 0.590786198879
LibB_ACCCAAACATGGCCTG-1 Sample0 1.0 0.00413587262409 1.0 0.000530469189207 0.0 0.00413587262409
LibB_TCAAGACCACCGATCG-1 Sample3 0.996511939525 0.00733825148285 0.00733825148285 0.00676639717093 0.0 0.996511939525
LibA_AGACAAAAGTAGACCG-1 Sample1 0.992696439997 0.00449414934583 0.0 0.992696439997 0.00449414934583 0.0
LibC_TAGGAGGGTGTCCTTC-1 Sample3 0.994666446983 0.0 0.0 0.0 0.0 0.994666446983
LibB_AGATTCGGTGATCAGG-1 Doublet 0.636443320426 0.345761737915 0.636443320426 0.345761737915 0.0 0.00109596204691
LibB_GAAGAGCGTGCTGGCT-1 Sample3 0.993330672446 0.0 0.0 0.0 0.0 0.993330672446
LibA_CCCTCTCAGGGCTCTC-1 Sample2 0.995806847294 0.0 0.0 0.0 0.995806847294 0.0
LibD_TAGTCCCAGATCTCAC-1 Sample0 0.994601202703 0.00139656759533 0.994601202703 0.0 0.0 0.00139656759533
We also generate a HTML file cotaining output figures
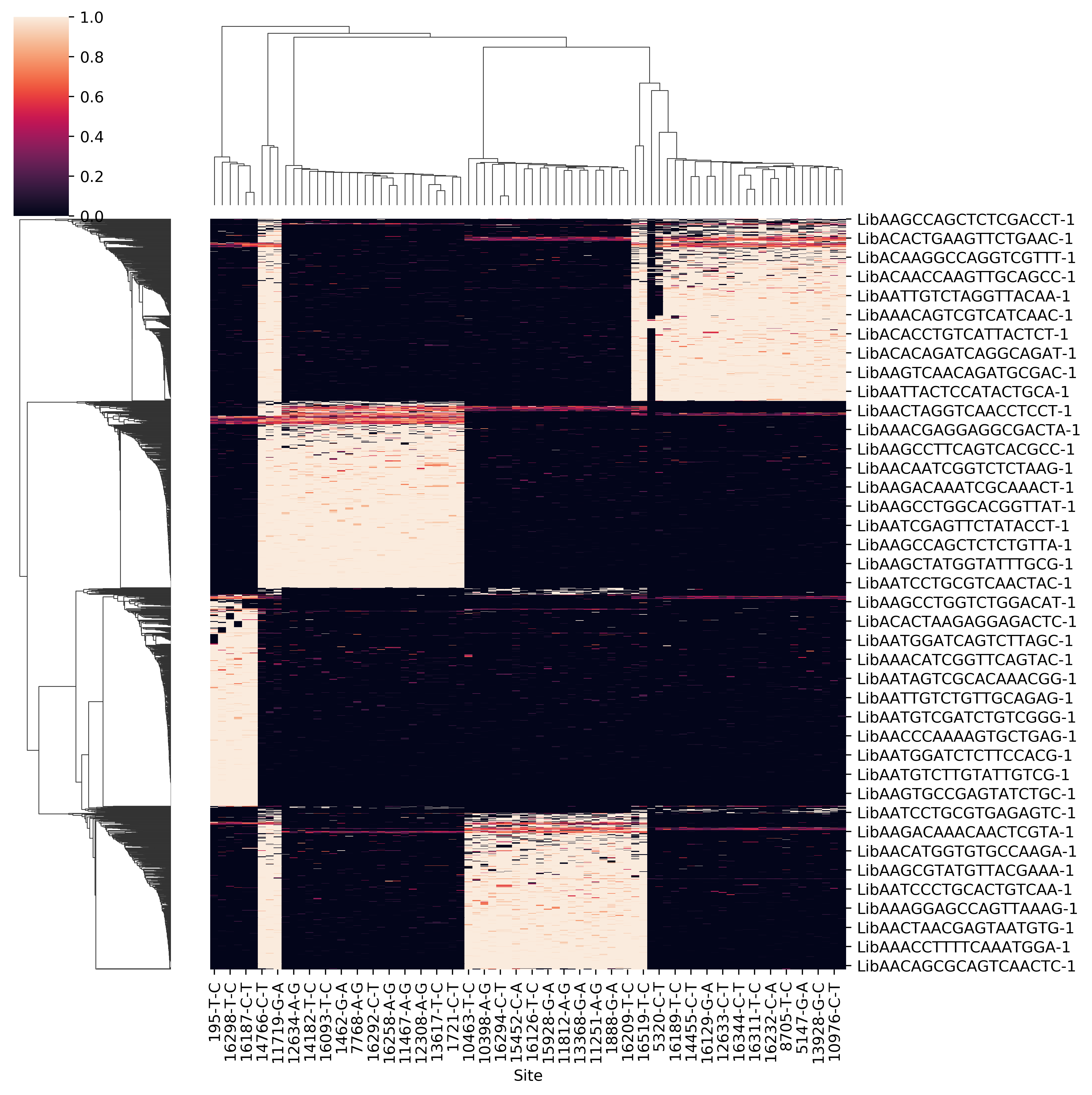
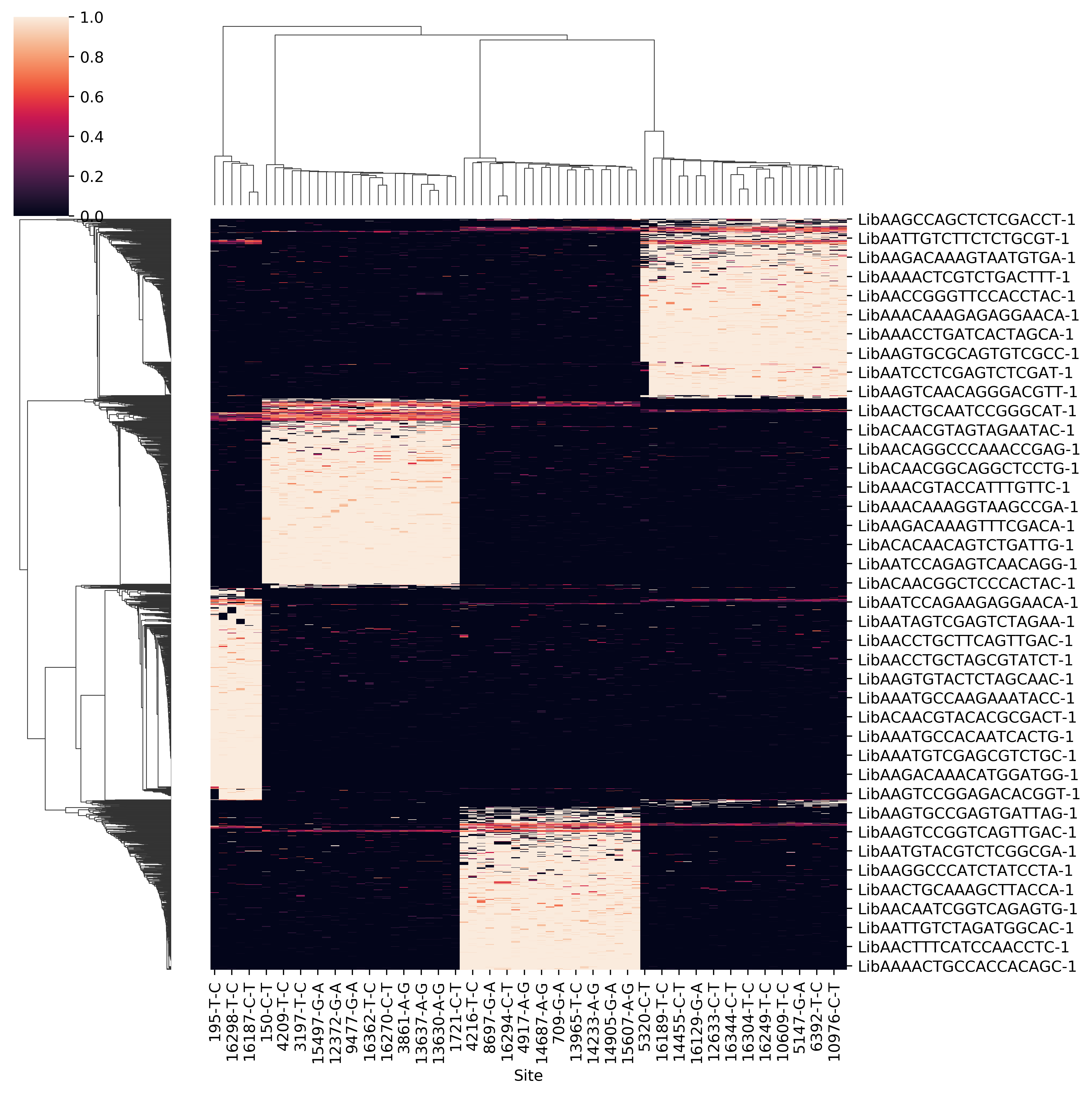
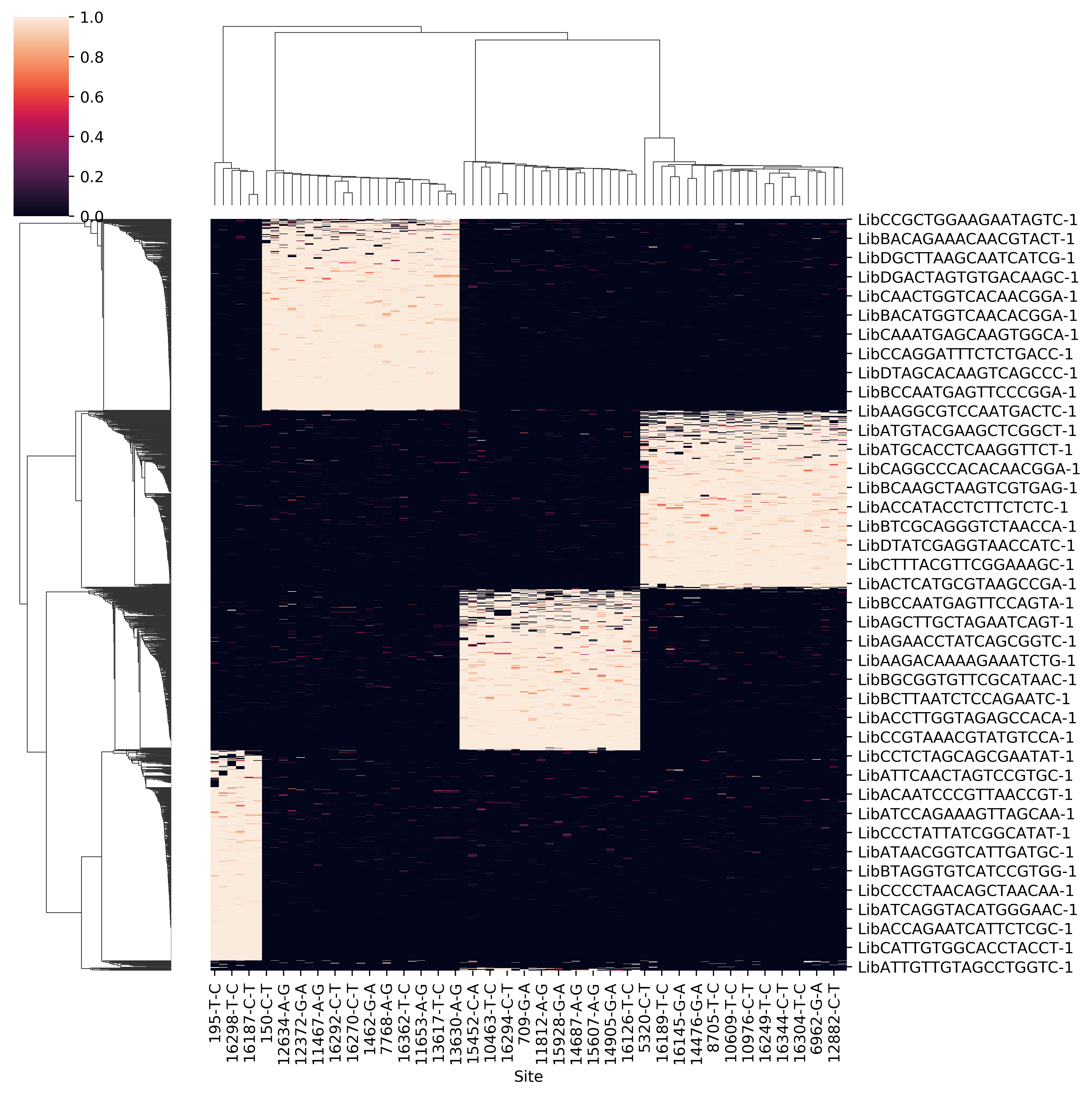
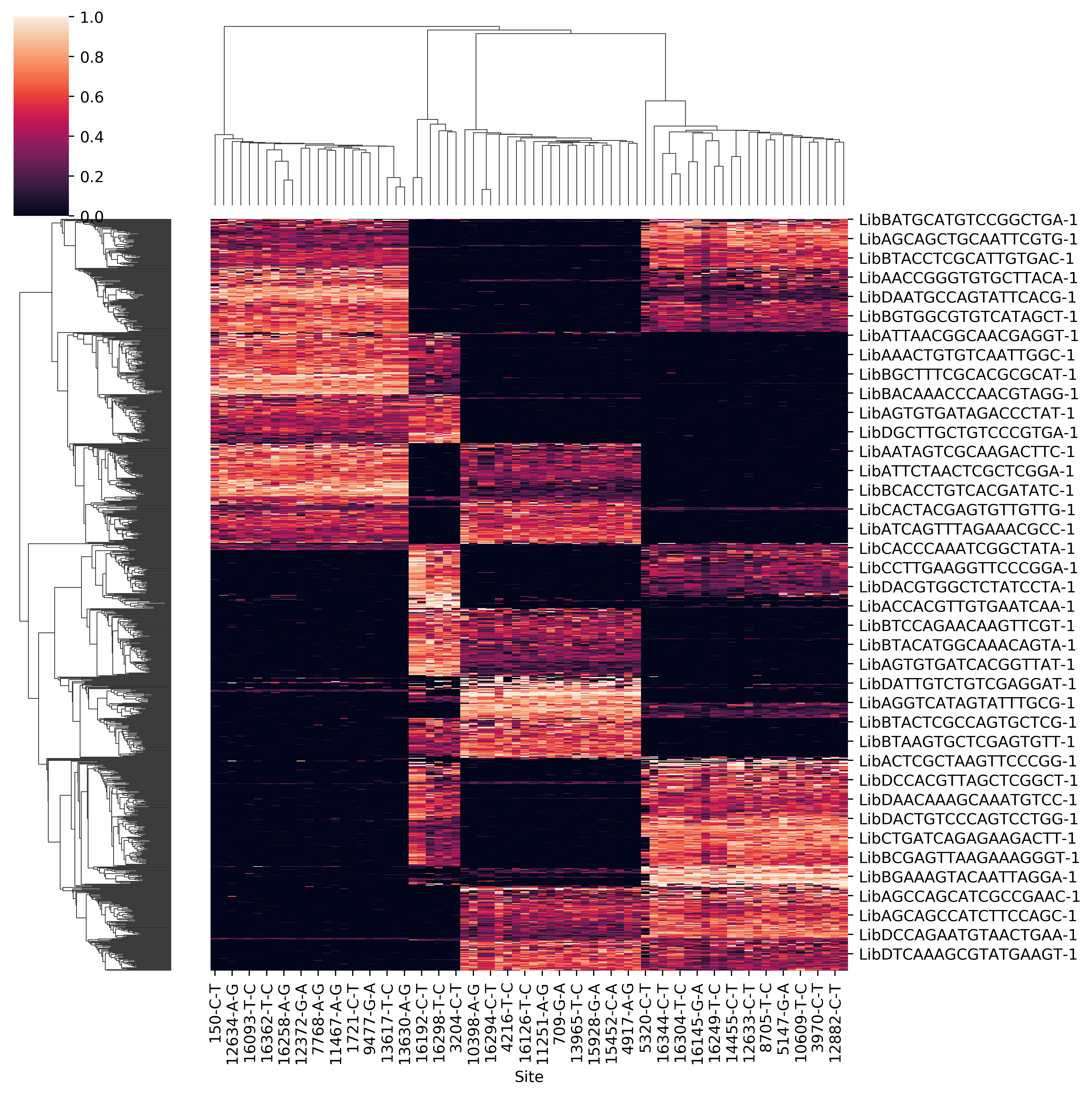
- Raw allele frequency matrix
- Clean allele frequency matrix for all cell barcodes (removing common variants)
- Scatter plot of p1 and p2 for doubelt identification. Users can modify the cutoff of p1 and p2 based on the plot
- Clean allele frequency matrix for singlets
- Clean allele frequency matrix for doublets
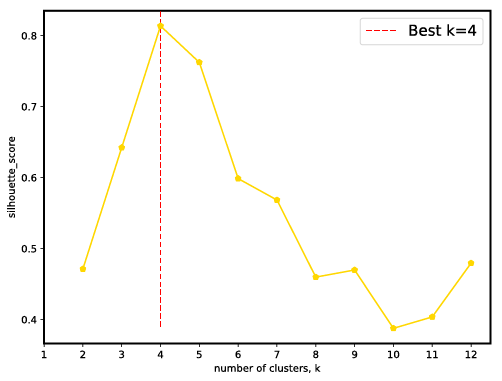
- Sihouette score for a range of k. Users can check if it match with known number of pooled samples. Besides, when the number of pooled samples is unkown, the parameter k can be set by hand according to sihouette score.
Please download testing data set, which contain downsampled scATAC-seq data from DOGMA-seq data (GSE200417) which multiplexed two donors :
- test_DOGMAseq_atac_possorted_chrM.bam
- test_DOGMAseq_atac_possorted_chrM.bam.bai
- test_DOGMAseq_barcode.txt
## clone MitoSort repository
git clone https://github.com/tangzhj/MitoSort.git
## create a conda environment and install all the required python packages
conda env create -f /path/to/MitoSort/MitoSort_env.yaml
conda activate MitoSort
## realign MT reads for test data, the reference genome is hg38
python /path/to/MitoSort/MitoSort_pipeline.py mt-realign -b /path/to/data/test_DOGMAseq_atac_possorted_chrM.bam -f /path/to/hg38.fasta --gatk_path /path/to/MitoSort/GenomeAnalysisTK_3.5-0.jar -o /path/to/output_dir
## generate SNP matrices
python /path/to/MitoSort/MitoSort_pipeline.py generate-snp-matrix -b /path/to/output_dir/MitoSort/BAM/possorted_chrM_realign.bam -f /path/to/hg38.fasta -c /path/to/data/test_DOGMAseq_barcode.txt -m /path/to/MitoSort/data/hg38_chrM.bed --varscan_path /path/to/MitoSort/VarScan.v2.3.7.jar -o /path/to/output_dir
## demultiplex samples
# you can tune the cutoff by checking the results of demultiplexing
python /path/to/MitoSort/MitoSort_pipeline.py demultiplex -o /path/to/output_dir -k 2 --p1_cutoff 0.8 --p2_cutoff 0.2