This is the code that got our team "Cappucino Monkeys" 4th place at The Allen AI Science Challenge Kaggle competition.
In this competition your program has to do well in standardized 8th grade science exam. The exam consists of multiple choice questions spanning biology, chemistry, physics, math and so on. Each question has four possible answers, of which exactly one is correct.
We ended up ensembling 5 deep learning models and 4 information retrieval models. Most of the code was written during one week at DeepHack.Q&A hackathon in Moscow.
Firstly the crucial part of our solution was the dataset. We collected 5M question-answer pairs from websites such as Studystack, Flashcardmachine and Quizlet. You can download part of the dataset below:
- Studystack (454743 questions)
Unpack the data to data folder. The same data is used both for training deep learning models and in information retrieval approach. Unfortunately the usage terms don't let us publish the Flashcardmachine and Quizlet portions.
You will also need training_set.tsv, validation_set.tsv and/or test_set.tsv provided by Allen Institute. Unfortunately these are not available for download any more, but those who participated in the competition probably have them. Copy them to data folder as well.
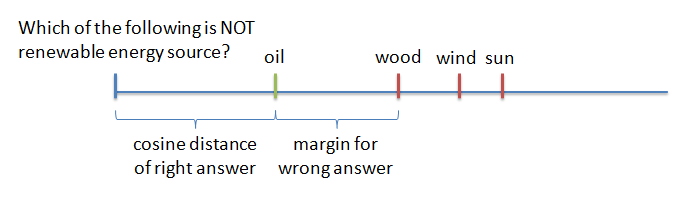
Our deep learning approach was inspired by a paper from IBM Watson group: LSTM-based Deep Learning Models for Non-factoid Answer Selection. We used recurrent neural networks to get sentence vector for both question and answer. Then we used cosine ranking loss to make cosine distance for the question and the right answer smaller than for the question and wrong answer (by some margin). In code we actually used cosine similarity but it is easier to explain using cosine distance.
One nice trick was, that we didn't have 3 separate (shared) branches in the network, but instead used one network and combined questions, right answers and wrong answers sequentially in batch. Loss function considered every third sample in batch as question, every other third as right answer and another third as wrong answer. This simplified the network architecture greatly and made it possible to use it in prediction phase with any number of answers.
Our dataset had only right answers, but loss function needed wrong answers as well, so we had to produce them ourselves. This is called "negative sampling" (right answer is the "positive sample" and wrong answer is the "negative sample"). We used strategy similar to Google FaceNet to choose wrong answers from the same (macro)batch.
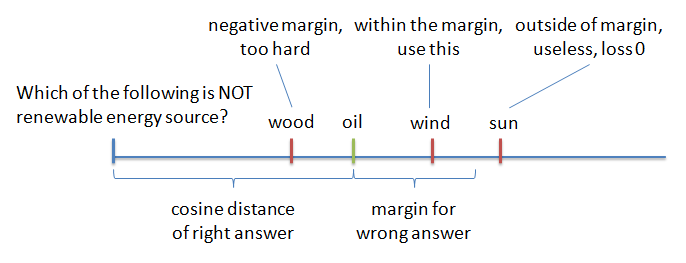
For the network to learn well, it is useful to not feed random wrong answers to it, but those that are "hard". That means answers that are wrong, but close to question in the cosine distance. But if you feed only the hardest questions to the network, it fails to converge, it is just "too hard". Instead people have found that using "semi-hard negative samples" - answers, which are further than the right answer, but still within the margin - works best. And that's what we did.
Another important feature was that after each epoch we tested our model on Allen AI training set and included the accuracy in the file name of saved weights. This allowed to track much easier how the currently trained models are doing and which can be killed to make room for subsequent experiments. At some point we had 10 GPUs running different experiments.
We used excellent Keras toolkit for implementing the model.
Follow Keras installation instructions.
To run the training with deep model:
python src/train_online.py model/test
The only required parameter is the path where to save the models to. In this example the file names will be something like model/test_00_loss_0.1960_acc_0.2868.hdf5, where 00 is the epoch number (starting from 0), 0.1960 is the training loss and 0.2868 is the accuracy on validation set (usually Allen AI training set).
Other parameters:
--data_path- path to data file. It should be text file with id, question and answer on each line, separated by tab.--csv_file- path to validation set, usually Allen AI training set file, which is default.--load_model- start training from pre-trained model.--load_tokenizer- the tokenizer to use, determines the number of words in vocabulary and embedding layer size. Default is Studystack full vocabulary tokenizer.--maxlen- maximum length of sentences (in words), default is 255.--macrobatch_size- size of macrobatch for negative sampling. Default is 1000.--min_margin- minimal margin for negative sampling, default is 0 (semi-hard).--max_margin- maximal margin for negative sampling, default is 0.2 (when margin is bigger than that, then gradient is 0 and the sample is basically useless for training).--rnn- eitherGRUorLSTM. Default is GRU, which seemed to converge faster in our experiments.--embed_size- size of embedding layer, default is 100.--hidden_size- size of hidden (RNN) layer, default is 512.--layers- number of hidden layers, default is 1.--dropout- dropout fraction (how much to drop), applied after every hidden layer. Default is 0, i.e. no dropout.--bidirectional-- use bidirectional RNN, works both with GRU and LSTM. Not enabled by default.--batch_size-- minibatch size, default is 300 (100 questions), must be multiple of 3.--margin-- margin to use in cosine ranking loss, default is 0.2.--optimizer- eitheradam,rmsproporsgd.adamis the default.--lr- learning rate to use. Works only when combined with--lr_epochs.--lr_epochs- halve learning rate after every this number of epochs.--samples_per_epoch- samples per epoch, default is 1500000 (500000 questions), should be multiple of 3.--epochs- maximum number of epochs.--patience- stop training when validation set accuracy hasn't improved for this number of epochs.
To run prediction with deep model:
python src/predict.py model/test_00_loss_0.1960_acc_0.2868.hdf5
The only required parameter is path to the model file. By default it just calculates accuracy on Allen AI training set, you need to use following options to produce predictions on other sets:
--csv_file- the file to calculate predictions for, default isdata/training_set.tsv, but can also bedata/validation_set.tsvordata/test_set.tsv. The code handles the missing "correct" column automatically.--write_predictions- write predictions in CSV file. The first column is question id, the second is predicted answer, followed by scores (cosine similarities) for all answers A, B, C and D. The file format was supposed to be compatible with Kaggle submission, but for ensembling purposes we had to include scores too.
For example to run prediction on test set:
python src/predict.py model/test_00_loss_0.1960_acc_0.2868.hdf5 --csv_file data/test_set.tsv --write_predictions results/deep_predictions_test.csv
Many options for the training script apply here as well. For example you need to match the hidden layer size and number of layers with saved model, otherwise it will give an error.
The preprocessing is mostly used to produce tokenizer. Tokenizer determines the vocabulary size and embedding layer size in the network. We used Keras default tokenizer class, which proved sufficient for our purposes. There is actually no need to run preprocessing, because the tokenizer for default dataset is already included in the repository. The instructions are included here just in case you want to use your own dataset.
First you need to get rid of the id field in original data file, so it doesn't clutter the vocabulary:
cut -f2,3 data/studystack_qa_cleaner_no_qm.txt >data/studystack.txt
Then run preprocessing on it:
python src/preprocess.py data/studystack.txt --save_tokenizer model/tokenizer_studystack.pkl
Additional options:
--max_words- limit the vocabulary to this number of most frequent words. Beware that if you limit the number of words, Keras default tokenizer just removes all the rare words from sentences, instead of replacing them with "UNKNOWN".
We did the information retrieval model on the last day and didn't tune it much. Basically we imported all our questions and answers to Lucene and used simple text search to find the most similar questions to given question and then matched the four possible answers with correct answers to those questions. Important idea was to multiply the scores of questions and answers, and sum all the results per each possible answer. This can be illustrated with following table:
| Question | Question Score | Answer A Score | Answer B Score | Answer C Score | Answer D Score |
|---|---|---|---|---|---|
| Question 1 | 6 | 0.1 | 0.1 | 0.4 | 0.1 |
| Question 2 | 4 | 0 | 0.2 | 0 | 0.1 |
| Question 3 | 2 | 0.3 | 0 | 0 | 0 |
| Total | 1.2 | 1.4 | 2.4 | 1.0 |
In the above example three most similar questions to given question are shown, with similarity scores 6, 4 and 2. Each of the four possible answers to given question are matched with the right answer for similar question, and scored accordingly. Scores of questions and answers are multiplied and summed. In this example answer C has the highest total score and is therefore the best bet for the right answer.
We used Lucene default scoring function both for scoring questions and answers. Because I couldn't find a way to calculate similarity of two strings in Lucene, I had to do it in really braindead way - I created a dummy index in memory, added only one document with answer there and then searched it with all possible answers. Surprisingly it wasn't that slow and worked rather well.
Using this approach with 10 most similar questions got us accuracy 42%. We were quite surprised, when using the exact same method with 100 most similar questions resulted in 49.6%! Using 1000 questions improved it further to 51.6%, but 2000 got us back to 50.88%. We are not sure yet why increasing the questions to ridiculous numbers works, but we noticed that in many cases all the answer scores are zeros, which always predicts answer A. Taking into account more questions might produce more non-zero scores for answers and give better chance for reasonable guess.
Run IPython notebook in lucene folder and go through the index.ipynb. This should produce folder index.
Open search.ipynb and step through it. You may want to change the number of similar questions on this line:
result = searcher.search(query, 100)
| Model | Training set | Preliminary test set | Final test set |
|---|---|---|---|
| Deep models | |||
| deep4264 | 42.64% | 41.437% | |
| deep4588 | 45.88% | 46.036% | |
| deep4704 | 47.04% | 45.466% | |
| deep4800 | 48.00% | 46.430% | |
| deep4812 | 48.12% | 46.605% | |
| Information retrieval models | |||
| lucene100 | 49.60% | 49.365% | |
| lucene500 | 51.16% | 50.591% | |
| lucene1000 | 51.60% | 48.125% | 49.803% |
| Google Search baseline | 42.17% | 45.204% | |
| Ensembles | |||
| Deep5+Lucene3+GS | *55.75% | 53.125% | 56.242% |
| Deep5 | *50.12% | 48.875% | 50.460% |
| Lucene3 | *51.76% | 49.750% | 51.205% |
| Deep5+Lucene3 | *54.95% | 50.750% | 54.621% |
(*) Ensemble model accuracies on training set are actually calculated on validation subset. Ensembling code is available on request.
Percentage of answers produced by different models that were the same:
| deep4264 | deep4588 | deep4704 | deep4800 | deep4812 | lucene100 | lucene500 | lucene1000 | GS | |
|---|---|---|---|---|---|---|---|---|---|
| deep4264 | 100.00% | ||||||||
| deep4588 | 44.93% | 100.00% | |||||||
| deep4704 | 43.60% | 55.31% | 100.00% | ||||||
| deep4800 | 45.33% | 61.09% | 65.53% | 100.00% | |||||
| deep4812 | 44.47% | 60.90% | 63.53% | 74.34% | 100.00% | ||||
| lucene100 | 35.70% | 39.11% | 38.04% | 39.06% | 38.99% | 100.00% | |||
| lucene500 | 37.38% | 40.81% | 39.97% | 40.60% | 40.70% | 69.12% | 100.00% | ||
| lucene1000 | 37.52% | 40.39% | 39.79% | 40.31% | 40.20% | 63.04% | 85.72% | 100.00% | |
| GS | 34.71% | 39.16% | 37.96% | 39.19% | 38.84% | 37.36% | 38.56% | 38.11% | 100.00% |
As can be seen, different models produce substantially different answers, even when the dataset used to train them was the same.
This was our first serious Kaggle competition and we never expected to be in the prize range. For that reason we did not upload our model and did not pay too much attention to the rules about which datasets and services can be used. But we hope we can serve the community by publishing our results with non-standard data sources as well.