Python library for pathology image analysis
This project is part of my PhD thesis to analyse histopathological images. Especially, the breast cancer hematoxylin and eosin-stained images from BreCaHAD: a dataset for breast cancer histopathological annotation and diagnosis
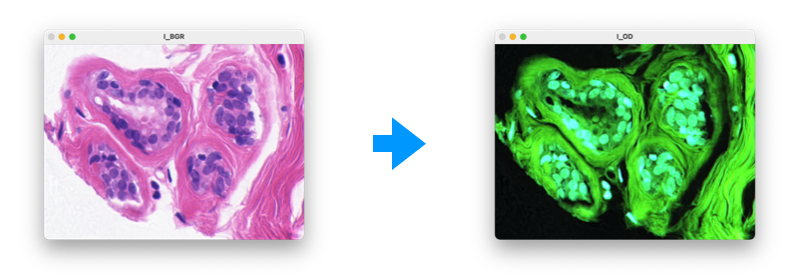
- Optical density transform
- Color deconvolution local_binary_patternslocal_binary_patterns
$ pip install python-patho$ python demo.py>>> from pypath.transform import convert_RGB_to_OD
>>> import cv2
>>> I_BGR = cv2.imread('images/he.png')
>>> I_RGB = cv2.cvtColor(I_BGR, cv2.COLOR_BGR2RGB)
>>> I_OD = convert_RGB_to_OD(I_RGB)>>> from pypath.stain_extractor import HE_color_decon
>>> I_H, I_E = HE_color_decon(I_RGB)>>> from pypath.texture import cal_lbp
>>> I_LBP = cal_lbp(I_BGR)>>> from pypath.utils import threshold
>>> from pypath.transform import mopho_process
>>>
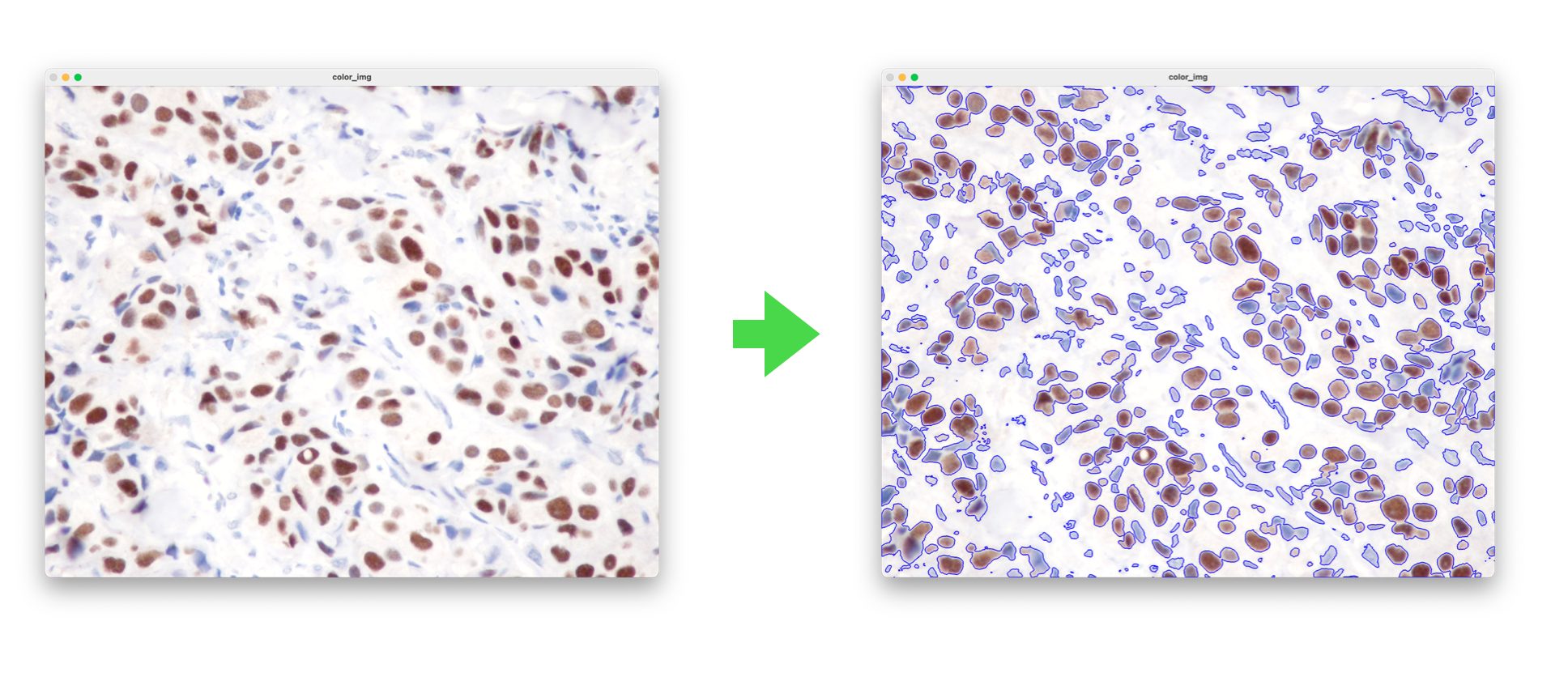
>>> src_path = 'IHC_IMAGE.jpg'
>>> color_img = cv2.imread(src_path)
>>> color_img = staintools.LuminosityStandardizer.standardize(color_img)
>>>
>>> gray = cv2.cvtColor(color_img, cv2.COLOR_BGR2GRAY)
>>> gray = threshold(gray, 220)
>>> cells_image = mopho_process(gray, gen_kernels())
>>>
>>> contours, hierarchy = cv2.findContours(cells_image, cv2.RETR_TREE, cv2.CHAIN_APPROX_NONE)
>>> cv2.drawContours(color_img, contours, -1, (255,0,0), 3)>>> from pypath.cell_processor import select_cell
>>> color_cell, binary_cell = select_cell(color_img, contours, 161)