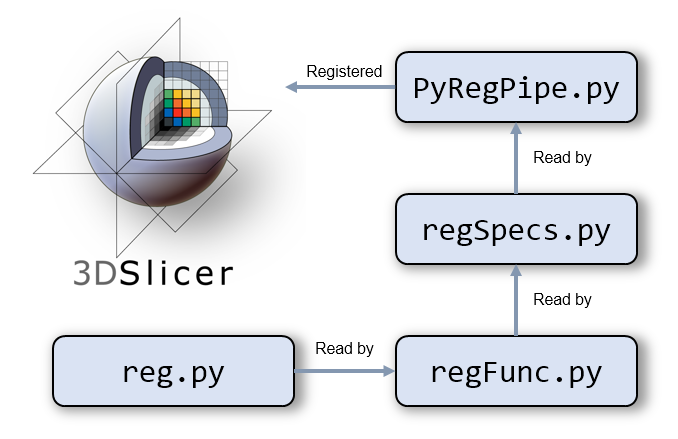
The automatic registration framework using Python for 3D Slicer, developed for the BiRT project.
Developed and maintained by Yu Sun. No Python objects are harmed during the development phase.
This framework utilises the Python environment in 3D Slicer for automating the registration steps developed by Reynolds, et al.
Reynolds, et. al., "Development of a Registration Framework to Validate MRI With Histology for Prostate Focal Therapy", Med Phys. 2015 Dec;42(12):7078-89. doi: 10.1118/1.4935343.
Versions at the time of creation:
- Python v3.6.7
- 3D Slicer v4.11.0
There are currently four modules (under the main folder):
PyRegPipe.py: the main module which implements the steps to perform in vivo to ex vivo registration.ToNIfTI.py: a module for converting DYNAMIKA DICOM to NIfTI filesPrepPk.py: a module for preparing the folders for DYNAMIKA pharmacokinetic maps.WarpImg.py: (for heritage files) a module for warping images from the in vivo space into the ex vivo space, given the exisitng CMTK transformation folder.WarpCnt.py: warp contours to the ex vivo space.GenSceneFile.py: generate the Slicer scene file after the registration finishes.
PyRegPipe is based on Python automation on 3D Slicer functions. The following text describes the background and the utilities which can be used elsewhere.
The PyRegPipe automates two parts:
- Conversion of in vivo data;
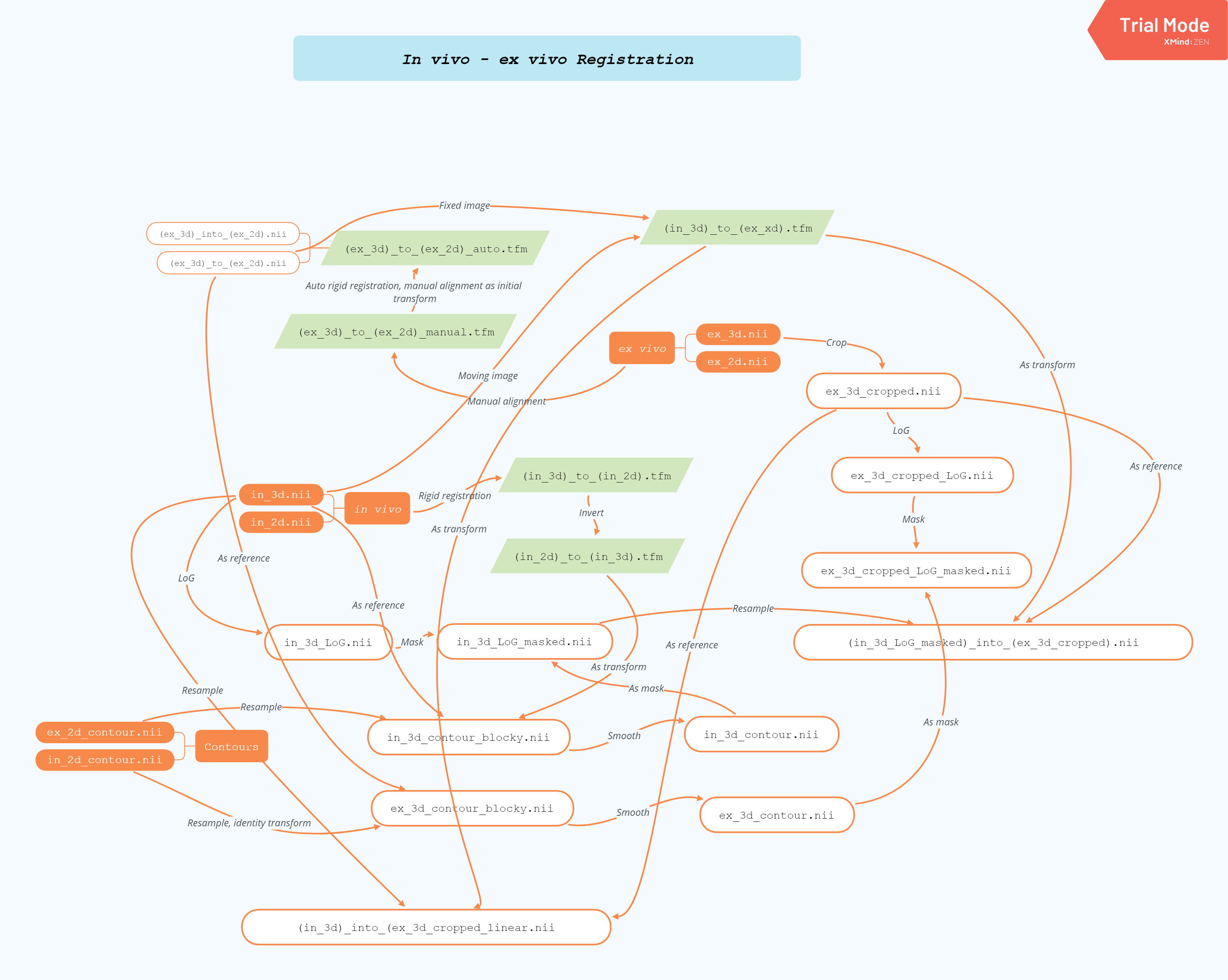
- Registration between in vivo and ex vivo data. This involves multiple steps which is shown in the diagram below.
Currently the framework only automates up to the in vivo - ex vivo registration. The histology to ex vivo registration still depends on MATLAB, so hasn't been included in the process yet.
There are two components regarding the file:
- A set of tools including 3D Slicer, CMTK, ImageJ and some bash utilities. These are in the folder
Tools; - The code which lives in the folder
code(this repository).
To set up and use the framework:
- Download the
PyRegPipe.ziphere and unzip into a folder (referred astopFolder). This contains theToolsfolder mentioned earlier and an emptycodefolder. - Download this repository and unzip into the
codefolder. This provides the modules for the registration framework.
This is the correct folder structure:
- topFolder
- Tools
- code
- cmtk
- main
- img
...
This is a portable version. You can put the everything on a USB stick.
Details are given in a step-by-step way on the BitBucket page.
The slice for motion correction is in dce_motion.py. Currently it's defined up to mrhist045. For new patients, you simply add the information at the end. Note that there is no comma after the last item.
{
...
'mrhist043': '16',
'mrhist044': '25',
'mrhist045': '15',
'new_patient': 'slice_number'
}
There are two major aspects for using Python in Slicer: automation and module development. First, Slicer has an embedded Python interpreter which can be used to automate most tasks. This includes loading a file, run a specific module, set the window content, etc. Secondly, Python can be used to develop what's called a scripted module in Slicer. These two aspects are often closely inter-related as the scripted module typically applies Python automation to achieve its goals.
Slicer has three types of modules, command line interface (CLI) modules, loadable modules and scripted modules. The last type can be developed using Python.
Slicer prior to version 4.11 embeds a Python 2 interpreter. From version 4.11 onward, it's Python 3. Since Python 3 is the standard (and not backward compatible with Python 2), we should stick to Python 3 as much as we could in general (even outside Slicer). At the time of writing this document (21/02/2020), the latest stable version of Slicer is 4.10.2 and the preview release version is 4.11.0.
The preview release (version 4.11.0) has greatly improved the integration of Python into Slicer. It is tailored for the vast Python user community in the field of medical imaging. Improvement includes the enhanced functionality for package management using pip along with the integration of the Python data science ecosystem (e.g. scipy and matplotlib). As a result, the development in our group is based on the preview release.
Click the Python icon to open the Python interactive shell, which will show up at the bottom of Slicer.
It is important to understand the architecture of Slicer so that you can write better scripts in the future. Slicer applies a special notation called Medical Reality Markup Language (MRML). As you will find out, most underlying classes in Slicer have the word "MRML" in their names. When Slicer is open, what you see is the MRML scene. This is similar to a stage where players can show up there. Inside the MRML scene, there are different MRML nodes. A node is similar to a player on stage. When you load a DICOM into Slicer, a scalar volume node is created. When you convert that into a label map, then it's converted into a label map node. It is possible that you create some node but don't include them in the scene, just as you may recruit some workers but they never show up on the stage. Nodes can also contains other nodes. For example, a scalar volume node may contain the storage node which stores the path of the original data.
Slicer has wrapped
SimpleITK,vtk,ctkandqt, which you can all access from the Python interpreter.
Slicer largely applies the ITK for image processing and VTK for visualisation. SimpleITK is the Python wrapper of the C++ implemented ITK. As mentioned in the sidebar, Slicer Python comes with SimpleITK which offers an extensive amount of image processing ability. Similarly, VTK is provided via the vtk module.
Qt is a versatile cross-platform graphical user interface (GUI) framework developed by Nokia. Slicer has wrapped the whole Qt (originally C++) in the module qt. Notice that qt is a Slicer-specific version of the C++ Qt, which is different from the more popular PyQt or PySide. Since there is no tkinter (the default GUI for Python on Windows) in Slicer, if you use matplotlib for plotting you need to set the back-end to qt.
In addition to qt, the Common Toolkit (ctk) provides some extra widgets you can use. The most common one is the dropdown list for selecting the input / output data in the CLI modules.
When you launch Slicer, you'll find a module called slicer. This is the main module you need to access the functionality of Slicer. The util sub-module contains the commonly used routines such as loading and saving data. The nodes (mentioned in the previous section) can be found in slicer, e.g. slicer.vtkMRMLScalarVolumeNode and slicer.vtkMRMLTransformNode.
The Python interface in Slicer offers a wide range of tools to automate tasks and create customised scripted modules. The current preview release (v4.11.0) ships with Python 3 which is the current standard. With slicer, SimpleITK, vtk, qt, ctk and other third-party libraries (e.g. 'Elastix'), this is probably the best environment one can find for medical image processing.
Next we will look at some examples.
This section includes some commonly used task to get you interested. More comprehensive examples can be found at Python Scripting and Script Repository.
>>> import sys
>>> sys.executable
'I:/Tools/Slicer 4.11.0-2019-11-25/bin/PythonSlicer.exe'It will be similar to a typical Python, but some modules related to the MRML are not loaded (e.g. slicer). So probably not that useful.
λ I:/Tools/"Slicer 4.11.0-2019-11-25"/bin/PythonSlicer.exe
Python 3.6.7 (default, Nov 25 2019, 23:26:12) [MSC v.1900 64 bit (AMD64)] on win32
Type "help", "copyright", "credits" or "license" for more information.
>>> import SimpleITK
>>> slicer
Traceback (most recent call last):
File "<stdin>", line 1, in <module>
NameError: name 'slicer' is not definedUse the pip_install() to install packages inside the Slicer Python.
>>> pip_install('pybrain')
Collecting pybrain
Downloading https://files.pythonhosted.org/packages/be/42/b40b64b7163d360425692db2f15a8c1d8fe4a18f1c5626cf7fcb5f5d5fb6/PyBrain-0.3.tar.gz (262kB)
Building wheels for collected packages: pybrain
Building wheel for pybrain (setup.py): started
Building wheel for pybrain (setup.py): finished with status 'done'
Created wheel for pybrain: filename=PyBrain-0.3-cp36-none-any.whl size=399048 sha256=eb62a5f0adb63f2ca87321849547527d680cc36b532f8af29198e80f2b5bd961
Stored in directory: C:\Users\ysun1030\AppData\Local\pip\Cache\wheels\21\60\92\907b453266e898b6914f7b175c7f346b4d7c349e2e28ce2b53
Successfully built pybrain
Installing collected WARNING: You are using pip version 19.3.1; however, version 20.0.2 is available.
You should consider upgrading via the 'python -m pip install --upgrade pip' command.
>>> packages: pybrain
Successfully installed pybrain-0.3To get the node (the underlying Slicer Python object), you need to know either the name or the ID of the node. The easiest way is to get the node by its name. This is the name shown under the Data module. For example, say we have loaded in_3d.nii and by default it will be shown as in_3d in Slicer. To get this node:
node = slicer.util.getNode('in_3d')Once you get the node, you can do a variety of operation on the node, similar to how you can process those shown under Data, e.g. thresholding the image, register with another image, put under a transformation.
Often you want to process the imaging data in a numpy array due to the rich utilities of numpy. Once you get the node, you can retrieve the pixel array as a numpy.ndarray.
>>> arr = slicer.util.arrayFromVolume(node)
>>> type(arr)
<class 'numpy.ndarray'>
>>> arr
array([[[ 24, 28, 31, ..., 4, 6, 0],
[ 21, 18, 29, ..., 11, 13, 0],
[ 7, 9, 10, ..., 29, 8, 0],
...,
[222, 233, 233, ..., 160, 155, 0],
[194, 254, 228, ..., 141, 139, 0],
[210, 244, 232, ..., 137, 135, 0]]], dtype=uint16)# Load a file and capture its returned node
in_3d = slicer.util.loadVolume(r'I:\reg\mrhist039\nifti\in_3d.nii')The resultant node will be returned. In previous version before 4.11.0, you need to pass returnNode=True to return the node. In version 4.11.0 onward, the node is by default returned.
The previous code is equivalent to
# Load the data, but not capture the node
slicer.util.loadVolume(r'I:\reg\mrhist039\nifti\in_3d.nii')
# Get the node, assuming the file is loaded as "in_3d" under the Data module
in_3d = slicer.util.getNode('in_3d')# Will return a Boolean to indicate save successfully or not
>>> slicer.util.saveNode(in_3d, r'D:/in_3d.nii')
True>>> slicer.util.loadScene(r'I:\reg\mrhist039\temp\11_manual_alignment.mrb')Notice this won't return any value, so you need to explicitly check whether it's successfully loaded.
>>> slicer.util.saveScene(r'I:\reg\mrhist039\temp\test.mrb')
TrueThis will return a Boolean value to indicate the success / failure .
A number of functions have been created for the basic tasks in the file reg.py. These functions are used in the registration pipeline. However, one can simply call any of these functions in isolation.
You can use these functions in the typical Python way: import the module and call the functions. The following code is supposed to be executed in the Python console within 3D Slicer.
To import the modules, add the path of code to the Python search path.
import sys
sys.path.append(r'I:/code')Then you can import reg.py since it's now under the search path. Here all functions are imported since it's unlikely to have name clashes with the system variables.
from reg import *You can call any function now. For example, to get the DICOM header attribute from a file:
getDcmAttr(r'I:/path/to/file.dcm', ('0019', '100c'))Please refer to the following section for the important functions.
This section shows the documentations of some of the important functions defined in reg.py. The file is located in the code folder.
getDcmAttr: get the header attribute in the DICOM file given the key.sortDcm: sort the DICOM files based on a specific key.cvtSlicer: convet an imaging file (specified by file extensions).cvtITK: convert a folder of DICOM files to another format (specified by file extensions using SimpleITK.rigidReg: rigid registration using BRAINS Registration.invertTfm: Invert an existing linear .tfm file.warpImg: resample an image using an exisiting transform.dceMotCorFolder: find the specific T1 subfolder for DYNAMIKA motion correction.copyHeader: copy the header information in the refFile to _file.dilateLabelMap: dilate a label map using Simple ITK.labelMapSmoothing: label map smoothing using slicer.cli.run().maskVolume: mask a volume using a label map.getParam: get the parameters of a Slicer CLI module.morphProcess: apply a morphological operation on a binary label map.logFilter: apply the LoG filter on an image.runCmd: run the command, return the ouptut and error.flipArr: flip an array (left-right, anterior-posterior and left-right).flipFile: read the image and save the flipped image.
With these building blocks, we will move to the design of the PyRegPipe.
The main goal is to speed up the iteration loop of the registration framework developed by Hayley. This is achieved by calling the functions developed in the reg.py. The framework won't improve the registration performance by itself, but will reduce the time for each step so each iteration loop can be faster.
In addition, the functions from reg.py can be used to define any customised workflow, which can greatly speed up the development of the biological atlas.
The framework is a module in 3D Slicer and complies with the requirements from Slicer. There are mainly three files involved, in addition to the underlying reg.py under the code folder. Technically speaking, all functions can be defined in one Python file. However, this is not the best practice as we want to separate the view from the logic.
-
PyRegPipe.py: this is the main module file which needs to be registered in Slicer, so that it will be loaded as a module. This file defines the meta-information, the GUI widgets and the logic (functions to be called). It will refer toregSpecs.pyto construct the main panel. -
regSpecs.py: this file contains the display of the main panel and links the buttons with the corresponding functions defined inregFunc.py. Hence when the user click a button, it will action accordingly. -
regFunc.py: this file defines the action of each step. It uses the functions defined in the utility filereg.py. The action for each step will be read byregSpecs.pyand linked with the buttons accessible to the user. -
reg.py: defines the commonly used tasks as functions. These functions can be called individually.
Let's have a closer look from the bottom up.
First, in regFunc.py, the path of the code folder is added to the Python search path. All functions in reg.py, along a few other separate functions, are imported.
# Add the code folder to the path
topFolder = os.path.abspath(os.path.join(sys.executable, *([os.pardir]*4)))
sys.path.append(os.path.join(topFolder, 'code'))
import genR2StarMacro
from genCmtkScript import genCmtkScript
from dce_motion import corrt_slice
from reg import *After that, there are functions that corresponds to each of the buttons in the automation panel. Each button and the corresponding function share a common name. For example, the first button (labeled as step1_1) is to convert the DICOM to NIfTI for in vivo 2D & 3D T2w images and rigidly register them. This corresponds to the function step1_1.
def step1_1(self):
'''Step 1: 1. T2w'''
key = 'step1_1'
output = self.mergeDict(
cvtITK(self.in_3d_dcm_folder, self.nii_folder, 'in_3d.nii'),
cvtITK(self.in_2d_dcm_folder, self.nii_folder, 'in_2d.nii'),
# Rigid registration, (in_2d)_to_(in_3d)
rigidReg(fixedImg=path.join(self.nii_folder, 'in_3d.nii'),
movingImg=path.join(self.nii_folder, 'in_2d.nii'),
outImg=path.join(self.nii_folder, '(in_2d)_into_(in_3d).nii'),
outTfm=path.join(self.tfm_folder, '(in_2d)_to_(in_3d).tfm')),
# Inverse the (in_2d)_to_(in_3d).tfm
invertTfm(path.join(self.tfm_folder, '(in_2d)_to_(in_3d).tfm'),
path.join(self.tfm_folder, '(in_3d)_to_(in_2d).tfm'))
)
update(self, key, output)This is similar to all other buttons. In total there are four tasks (if BOLD data is present, otherwise three tasks) in Step 1 and five tasks in Step 2.
Moving onto regSpecs.py, regFunc is firstly imported so that it has access to all functions in regFunc.py.
import regFuncNext, there is a dictionary specs which specifies the display of each task and links it with the corresponding function. For example, step1_1:
step1_1 = dict(
step = 'step1',
key = 'step1_1',
stepLabel = '''Tasks:
# 1. Convert in_2d & in_3d to NIfTI;
# 2. Coregister in_2d to in_3d;
# 3. Reverse the transformation file;
''',
btnFunc = regFunc.step1_1,
title = 'T2w Images',
shortDesc='T2w Images'
)In PyRegPipe.py, regSpecs.py is firstly imported.
import regSpecsThen there is a function which reads the content from regSepcs and create the corresponding GUI widgets.
def createSession(self, step, key, stepLabel, btnFunc, title, shortDesc):
layout = self.formFramDict[step].layout()
boxLayout = qt.QHBoxLayout()
*ignore, fFrame = self.createClp(title, layout, boxLayout)
self.sessFrames[key] = fFrame
self.createLabel(shortDesc, fFrame.layout(), stepLabel)
self.createBtn('Apply', lambda:btnFunc(self), fFrame.layout(), shortDesc)
self.statusLight[key] = self.createLabel(' ', fFrame.layout())
self.statusLight[key].setMaximumWidth(20)
self.setStatus(key, 0)Then the content of the regSpec is iterated for creating the widgets.
keys = list(regSpecs.specs.keys())
# If no BOLD, then skip step1_3
if not hasBold:
keys.pop(keys.index('step1_3'))
for key in keys:
self.createSession(**regSpecs.specs[key])A project file need to be specified. The automated framework will manage the files in different sub-folders:
Data: this is the only folder that needs to be set up by the user. It needs to have the following structure. Except forprostate_contours_from_slicer, the other folders will contain the DICOM images. Once the user specifies the project folder, the automated framework will check the existence ofdataand its structure.
- data
- in_vivo_mri
- in_3d
- in_2d
- dwi
- adc
- bold (optional)
- dynamic
- pk
- iauc
- ire
- ...
- ex_vivo_mri
- ex_2d
- ex_3d
- prostate_contours_from_slicer
- xx_in_2d-label.nrrd (xx is the patient number)
- xx_ex_2d-label.nrrd (xx is the patient number)
nifti: this folder contains the NIfTI (.nii) files generated.transformation: contains the transformation (.tfm) files generated.sorted: stores any sorted DICOM images, e.g. the BOLD data sorted by TE.script: stores the scripts generated, e.g. the ImageJ script for R2* map calculation.temp: stores any temporary data, e.g. the.mhdfiles for copying header information.cmtk: contains the script and initial files to be run by CMTK.
The Python automated framework GUI also contains:
- A file list for viewing the files stored in the
niftiandtransformationfolder; - A header copier for copying header information;
- A image flipper for flipping the image in one of left-right, superior-inferior, anterior-posterior. Header information will be maintained.
This is the end of the document.