Autoencoders - a deep neural network was used for feature extraction followed by clustering of the "Cancer" dataset using k-means technique
This project is an attempt to use “Autoencoders” which is a non-linear dimensionality reduction technique for feature extraction and then use the hidden layer activations which is given as input to the k-means algorithm for clustering. This project has two main components:-
Autoencoders : In this module, the objective is to give the .csv file as input to the input layer, get the hidden layer activations from the hidden layer. This is done using the gradient descent algorithm. The loss function used is the cross entropy loss function. The hidden layer activations are given as input to kmeans algorithm for clustering.
-
K-means : Linearly clustering the input where the input comes from the autoencoders and displaying the confusion matrix and clustering accuracy.
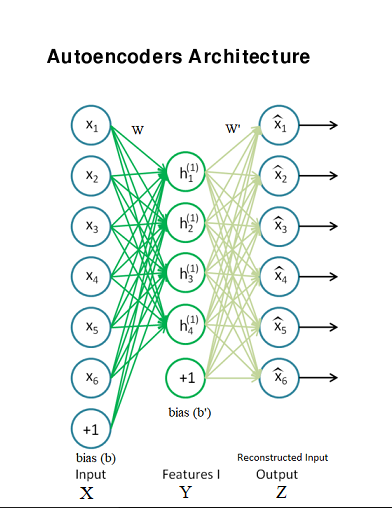
Autoencoders
Input : Input data matrix, No of hidden neurons, Weight matrix(W), No of clusters for k-means.
Let :
• X is the input data
• Y is the hidden layer activations
• Z is the predicted output or the reconstruction of the input X.
• W denote the weights from input to hidden layer
• b is the input and hidden layer bias
• s(.) denote the sigmoidal function
-
Take the input X ε [0,1] and map it ( with an encoder ) to a hidden representation y ε [0,1] through a deterministic mapping.
-
The latent representation , or code is then mapped back (with a decoder) into a reconstruction of the same shape as . The mapping happens through a similar transformation.
-
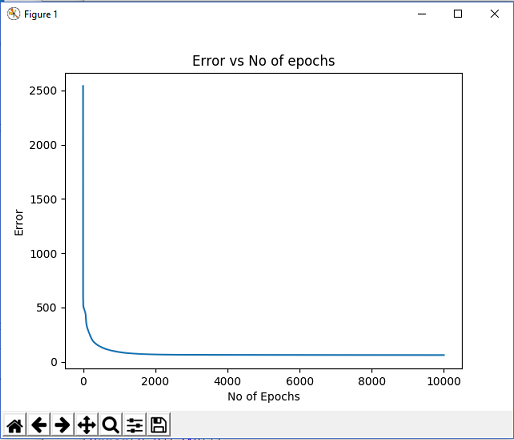
The reconstruction error is calculated using the cross- entropy loss function.
-
The weights are updated using the gradient descent equation.
**K-means Clustering : **
- Initialize the centroids randomly.
- Update the centroids based on the Eucledian distance.
- Group the datapoints based on minimum distance.
- Perform steps 5,6,7 for a certain number of iterations.
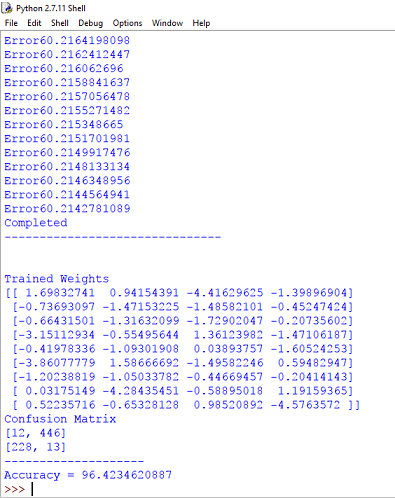
Output : Confusion Matrix and Clustering Accuracy
[1] P. Vincent, H. Larochelle, Y. Bengio, P.A. Manzagol: Extracting and
Composing Robust Features with Denoising Autoencoders, ICML'08, 1096-1103,
2008
[2] Y. Bengio, P. Lamblin, D. Popovici, H. Larochelle: Greedy Layer-Wise
Training of Deep Networks, Advances in Neural Information Processing
Systems 19, 2007
[3] https://github.com/lisa-lab