This repo is the official implementation for paper:
Meta-Polyp: a baseline for efficient Polyp segmentation.
Author: Quoc-Huy Trinh
In the IEEE 36th International Symposium on Computer Based Medical Systems (CBMS) 2023.
Detail of each model modules can be found in original paper. Please citation if you use our implementation for research purpose.
-
Upcomming: Supervision version
-
30/05/2023: Our paper "Meta-former UNet: Simple but effcient" is accepted EUSIPCO 2023. This is the specific concept of the Meta-former with UNet
-
19/05/2023: Update pretrained-weight
-
16/05/2023: Update paper link
-
13/05/2023: First commit
-
03/05/2023: Our paper "Meta-Polyp: a baseline for efficient Polyp segmentation" was accepted CBMS 2023.
Architecutre Meta-Polyp baseline model:
Our implementation is on Python 3.9 , please make sure to config your environment compatible with the requirements.
To install all packages, use requirements.txt file to install. Install with pip by the following command:
pip install -r requirements.txt
All packages will be automatically installed.
All of configs for training and benchmark are in ./config/ folder. Please take a look for tuning phase.
For training, use train.py file for start training.
The following command should be used:
python train.py
For benchmar, use benchmark.py file for start testing.
The following command should be used:
python benchmark.py
Note: You should fix model_path for your model path and directory to your benchmark dataset.
The pretrained weight for the MetaPolyp can found at Google Drive.
Note: For further usage, if are there any problems, please contact me via email from the paper, I am willing to support
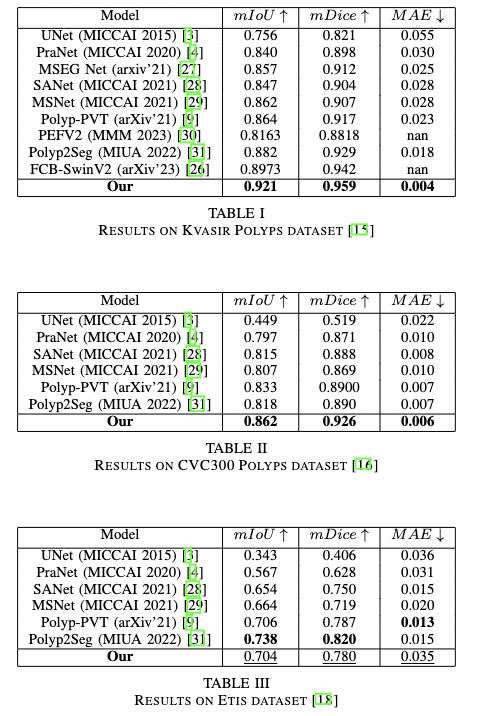
In our experiment, we use the dataset config from PraNet, with training set from 50% of Kvasir-SEG and 50% of ClinicDB dataset.
With our test dataset, we use the following:
In same distribution:
-
Kvasir SEG
-
ClinicDB
Out of distribution:
-
Etis dataset
-
ColonDB
-
CVC300
The IOU score on SOTA for both 5 datasets:
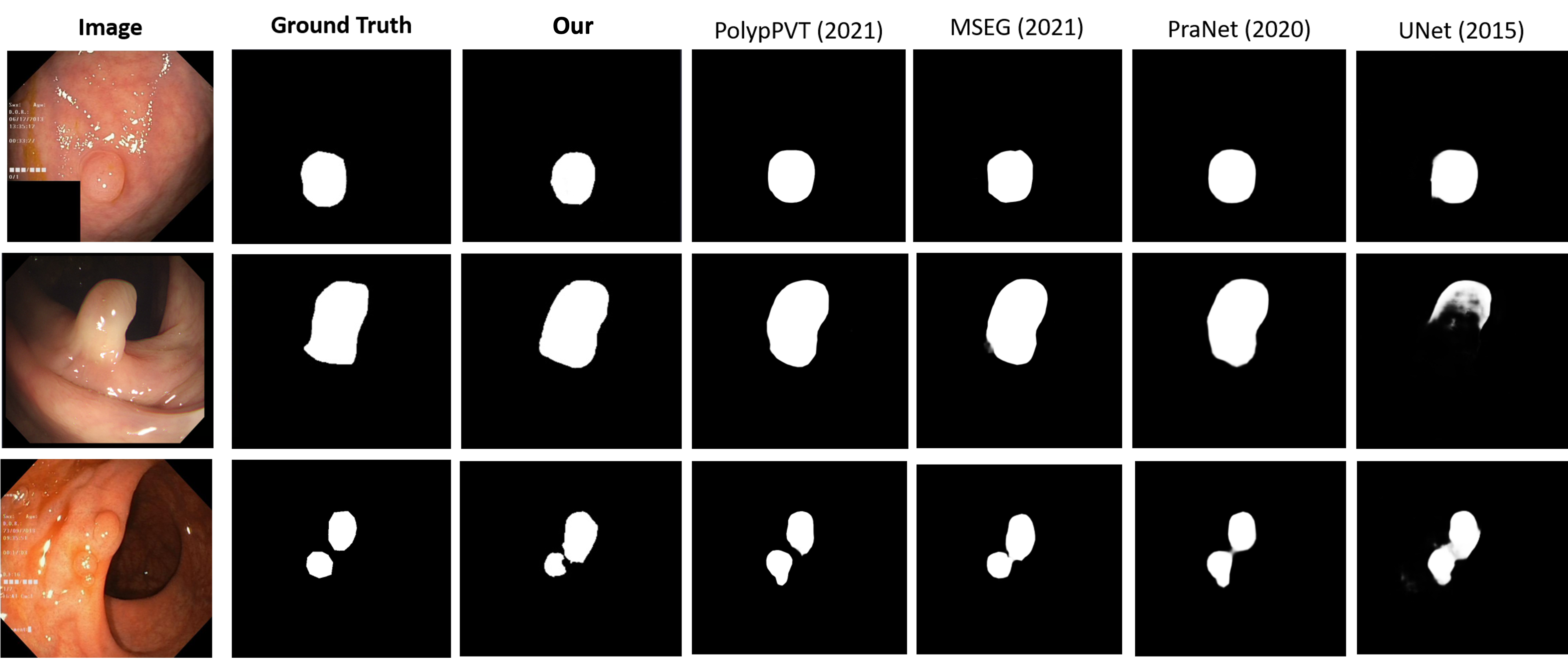
We do some qualiative result with others SOTA method visualization:
Coming soon
You can change the backbone from Ca-former to PVT or something else to get different results.
@misc{trinh2023metapolyp,
title={Meta-Polyp: a baseline for efficient Polyp segmentation},
author={Quoc-Huy Trinh},
year={2023},
eprint={2305.07848},
archivePrefix={arXiv},
primaryClass={eess.IV}
}