A set of tools to evaluate the reproducibility of computations.
verifyFiles.py compares the output files produced by pipelines in different conditions. It identifies the files that are common to all conditions, and for these files, it compares them based on their checksums and other metrics configurable by file type.
Python 2.7.5
usage:verifyFiles.py result_base_name [-h] [-c CHECKSUMFILE] [-d FILEDIFF] [-m METRICSFILE] [-e EXCLUDEITEMS] [-k CHECKCORRUPTION] [-s SQLITEFILE] [-x EXECFILE] [-t TRACKPROCESSES]
[-i FILEWISEMETRICVALUE] file_in
file_in, Mandatory parameter.Directory path to the file containing the conditions.
results_base_name, Base name to use in output file names.
-c,CHECKSUM FILE, --checksumFile Reads checksum from files. Doesn't compute checksums locally if this parameter is set.
-m METRICSFILE, --metricsFile CSV file containing metrics definition. Every line contains 4 elements: metric_name,file_extension,command_to_run,output_file_name
-e EXCLUDEITEMS, --excludeItems The path to the file containing the folders and files that should be excluded from creating checksums.
-k CHECKCORRUPTION, --checkCorruption The script verifies if any files are corrupted ,when this flag is set as true
-s SQLITEFILE, --sqLiteFile The path to the SQLITE file,having the reprozip trace details.
-x EXECFILE , --execFile Writes the executable details to a file.
-t TRACK PROCESSES, --trackProcesses If this flag is set, it traces all the processes using reprozip to record the details and writes it into a csv with with the given name
-i FILEWISEMETRICVALUE, --filewiseMetricValue Folder name on to which the individual filewise metric values are written to a csv file
Pytest syntax
pytest --cov=./ ./test_verifyFiles.py
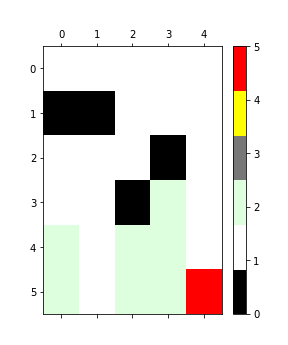
plot_matrix.py plot heatmaps of difference matrices produced by
verifyFiles.py. For instance, ./plot_matrix.py test/test_differences_plot.txt output.png will produce the following
plot:
-t argument gives the possibility to superimpose the predicted matrices achived by predict.py over the difference matrices produced by verifyFiles.py. For example, python plot_matrix.py test/predict_test/test_differences.txt -t test/predict_test/triangular-S_0.6_test_data_matrix.txt test_plot_matrix.png will make the following plot:
The aim of PEDS is clustering processes and creating a graph model representation for the all processes which introduce errors in the pipeline
based on the reprozip trace file (trace.sqlite3) and matrix-file created by veryfyFiles.py that specify the pipeline output files with defferences.
Python 2.7.5, graphviz module
-
peds.py [-db: database file from reprozip trace] [-ofile: the matrix-file from veryfyFiles.py]*Note: The output would be a dot format file which requiers to create appropriate representing file formats such as: svg, png and etc.
-
dot -Tpng Graphmodel.dot -o Figure.png
predict.py can be used to predict the elements of utility matrix M ij when following the sequential generating of elements is a concern.
(Ex. a comparison matrix of generated files from a same pipeline process in two different versions of an operating system)
The sampling method options for fitting the training sets of the Alternating Least Square (ALS) are consist of:
- columns
- rows,random-real
- random-unreal
- diagonal (random picking of j from a uniform distribution)
- triangular-L (Random-triangle-L: fewer i, more j)
- triangular-S (Random-triangular-S: more i, fewer j)
- Bias
Spark 2.2.0, Python 2.7.13
predict.py [utility matrix file][training ratio][training sampling method]