-
Notifications
You must be signed in to change notification settings - Fork 98
Example: Neural network WSI classification
Erik Smistad edited this page Aug 6, 2019
·
1 revision
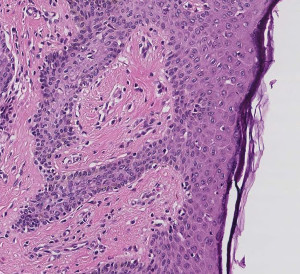
This example performs patch-wise classification of a whole slide microscopy image using a neural network.
/**
* Examples/Segmentation/neuralNetworkWSISegmentation.cpp
*
* If you edit this example, please also update the wiki and source code file in the repository.
*/
#include <FAST/Tools/CommandLineParser.hpp>
#include <FAST/Algorithms/NeuralNetwork/InferenceEngineManager.hpp>
#include <FAST/Visualization/SimpleWindow.hpp>
#include <FAST/Visualization/HeatmapRenderer/HeatmapRenderer.hpp>
#include <FAST/Importers/WholeSlideImageImporter.hpp>
#include <FAST/Visualization/ImagePyramidRenderer/ImagePyramidRenderer.hpp>
#include <FAST/Algorithms/ImagePatch/PatchGenerator.hpp>
#include <FAST/Algorithms/ImagePatch/PatchStitcher.hpp>
#include <FAST/Algorithms/NeuralNetwork/SegmentationNetwork.hpp>
#include <FAST/Algorithms/TissueSegmentation/TissueSegmentation.hpp>
using namespace fast;
int main(int argc, char** argv) {
Reporter::setGlobalReportMethod(Reporter::COUT);
CommandLineParser parser("Neural network WSI classification example");
parser.addChoice("inference-engine",
{"default", "OpenVINO", "TensorFlowCPU", "TensorFlowCUDA", "TensorRT", "TensorFlowROCm"},
"default",
"Which neural network inference engine to use");
parser.addPositionVariable(1,
"filename",
Config::getTestDataPath() + "/WSI/A05.svs",
"WSI to process");
parser.parse(argc, argv);
auto importer = WholeSlideImageImporter::New();
importer->setFilename(parser.get("filename"));
auto tissueSegmentation = TissueSegmentation::New();
tissueSegmentation->setInputConnection(importer->getOutputPort());
auto generator = PatchGenerator::New();
generator->setPatchSize(512, 512);
generator->setPatchLevel(0);
generator->setInputConnection(importer->getOutputPort());
generator->setInputConnection(1, tissueSegmentation->getOutputPort());
auto network = NeuralNetwork::New();
if(parser.get("inference-engine") != "default") {
network->setInferenceEngine(parser.get("inference-engine"));
}
const auto engine = network->getInferenceEngine()->getName();
network->setInferenceEngine(engine);
if(engine.substr(0, 10) == "TensorFlow") {
network->setOutputNode(0, "sequential/dense_1/Softmax", NodeType::TENSOR);
} else if(engine == "TensorRT") {
network->setInputNode(0, "input_1", NodeType::IMAGE, TensorShape{-1, 3, 512, 512});
network->setOutputNode(0, "sequential/dense_1/Softmax", NodeType::TENSOR, TensorShape{-1, 3});
}
network->load(Config::getTestDataPath() + "NeuralNetworkModels/wsi_classification." +
network->getInferenceEngine()->getDefaultFileExtension());
network->setInputConnection(generator->getOutputPort());
network->setScaleFactor(1.0f / 255.0f);
auto stitcher = PatchStitcher::New();
stitcher->setInputConnection(network->getOutputPort());
auto renderer = ImagePyramidRenderer::New();
renderer->addInputConnection(importer->getOutputPort());
auto heatmapRenderer = HeatmapRenderer::New();
heatmapRenderer->addInputConnection(stitcher->getOutputPort());
//heatmapRenderer->setMinConfidence(0.5);
heatmapRenderer->setMaxOpacity(0.3);
auto window = SimpleWindow::New();
window->addRenderer(renderer);
window->addRenderer(heatmapRenderer);
window->enableMaximized();
window->set2DMode();
window->start();
}If this wiki page lacks some information or is incorrect please let us know! You can edit this wiki page yourself, send an email to [email protected] or