This is a package for aiding in studying SDEs.
You can install the package via from github on windows/mac in command line with:
python -m pip install git+https://github.com/shill1729/sdepy.git
The package has functions/classes for
- Infinitesimal generators
- Adjoint generators
- SDE coefficients for Riemannian Brownian motion in local coordinates
- SDE coefficients for Riemannian Brownian motion in embedded in
$\mathbb{R}^D$
- Euler-Maruyama
- Milstein (TODO)
- Runge-Kutta (TODO)
- Monte-Carlo parabolic PDE solvers (via Feynman-Kac formula)
- Implicit finite difference solvers for parabolic PDEs
- Implicit finite difference solver for the telegrapher type
- Monte-Carlo solvers for the telegrapher PDE
- The Euler-Maruyama scheme gives a discrete-time Gaussian-Markov process approximation to the SDE
- We can parameterize the SDE coefficients as neural networks and then maximize this approximate likelihood
- Milstein-scheme leads to another discrete-time Markov process but is no longer simply Gaussian (TODO)
The user can supply coordinates
import matplotlib.pyplot as plt
from sdepy.symsde import *
from sdepy.solvers import euler_maruyama
# sympy input
t, x, y = sp.symbols("t x y", real=True)
xi = sp.Matrix([x, y])
coord = sp.Matrix([x, y])
g = metric_tensor(xi, coord)
# Path input
x0 = np.array([1.5, 3.14])
tn = 1
seed = 17
n = 9000
mu, sigma = local_bm_coefficients(g, coord)
f = sp.Function("f")(*coord)
inf_gen = infinitesimal_generator(f, coord, mu, sigma)
adj_gen = adjoint_generator(f, coord, mu, sigma)
print("Metric tensor")
print(g)
print("Local drift")
print(mu)
print("Local diffusion")
print(sigma)
print("Infinitesimal generator")
print(inf_gen)
print("Fokker Planck RHS")
print(adj_gen)
# Do any substitutions here
mu_np, sigma_np = sympy_to_numpy_coefficients(mu, sigma, coord)
xt = euler_maruyama(x0, tn, mu_np, sigma_np, n, seed=seed)
fig = plt.figure()
ax = plt.subplot(111)
ax.plot3D(xt[:, 0], xt[:, 1], color="black")
plt.show()Many every day surfaces can be written as the zeros of some smooth function:
- Sphere:
$f(x,y,z)=x^2+y^2+z^2-1$ - Ellipsoid
$f(x,y,z)=(x/a)^2+(y/b)^2+(z/c)^2-1$ - Paraboloid:
$f(x,y,z)=(x/a)^2+(y/b)^2-z$ - Hyperbolic Paraboloid:
$f(x,y,z)=(y/b)^2-(x/a)^2-z$ - Hyperboloid
$f(x,y,z)=(x/a)^2+(y/b)^2-(z/c)^2-1$ - Cylinder
$f(x,y,z)=x^2+y^2-1$ - Torus
$f(x,y,z)=(\sqrt{x^2+y^2}-R)^2+z^2-r^2$
The orthogonal projection method to generate Brownian motion is as follows. Given a surface
Then the Stratonovich SDE for BM on
More generally, for intersections of hypersurfaces we have
The function
For solutions to the SDE to exist, we only require locally Lipschitz continuity of
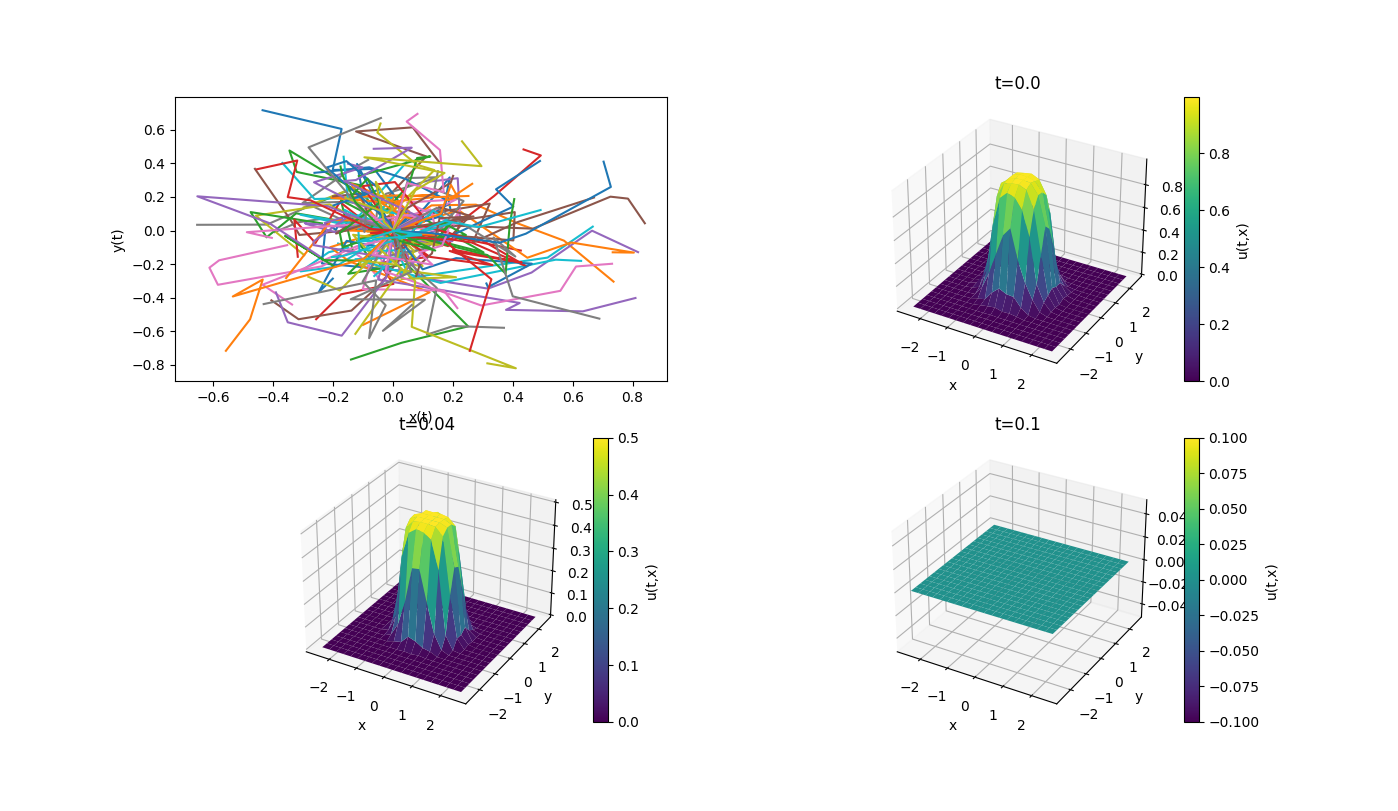
The below example computes the proportion of time that planar Brownian motion
spends in the unit-disk, i.e. we compute
In terms of the above notation, the terminal cost is
# A template for 2d Fenyman-Kac problems (solving PDEs with MC estimates of SDEs)
from sdepy.sdes import SDE
import numpy as np
tn = 0.1
ntime = 5
npaths = 50
noise_dim = None
x0 = np.array([1., 1.])
a = -1.5
b = 1.5
c = -1.5
d = 1.5
space_grid_size = 20
time_grid_size = 5
grid_bds = [a, b, c, d]
grid_sizes = [space_grid_size, time_grid_size]
def mu(t, x):
return np.zeros(2)
def sigma(t, x):
return np.eye(2)
def f(t, x):
# return np.abs(x) < 1.
return (np.linalg.norm(x, axis=1) < 1.)/tn
def h(x):
return 0.
# For 2-d PDE estimation
sde = SDE(mu, sigma)
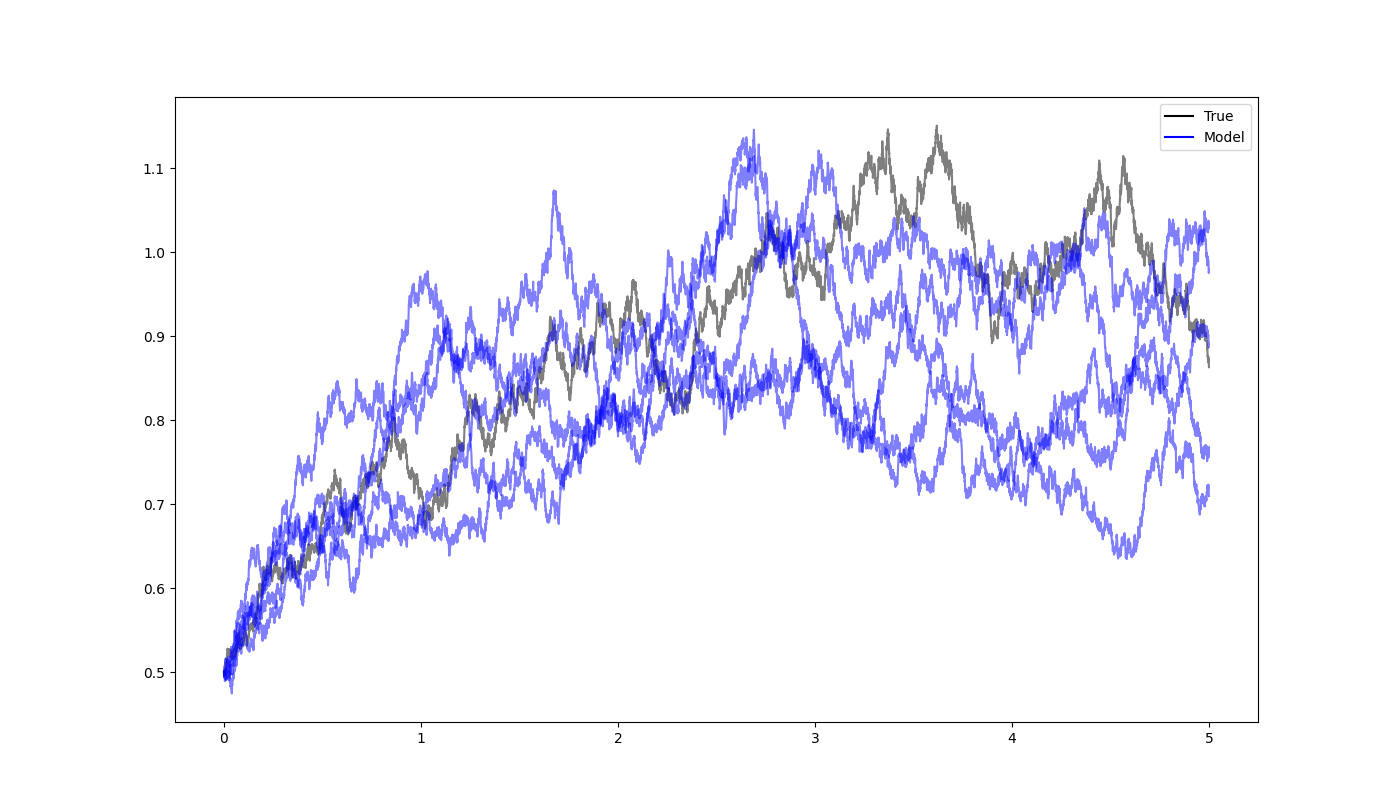
sde.feynman_kac_2d(f, h, x0, tn, grid_bds, grid_sizes, ntime, npaths, noise_dim)Every stochastic integration scheme yields a discrete-time, finite-order, Markov process approximation to the original SDE. The conditional distribution of this approximate process will have parameters in terms of the original SDE coefficient functions. If we replace these with neural networks, then we can approximate the original one by maximizing the (log)-likelihood of this approximate process over the neural-network parameters (weights and biases).
For example, the Euler-Maruyama scheme for our SDE says that
Recall that for any Markov process
Thus we can minimize the negative log-likelihood as our loss function and this will give an MLE for our neural SDE
model given an ensemble data-set with equal time-steps. The first two terms do not depend on the network parameters
We simulate a true CIR process over
# An example of our Neural SDE being fit to planar motions.
import matplotlib.pyplot as plt
import numpy as np
import torch
from sdepy.sdes import SDE
from sdepy.neural_sdes import NeuralSDE
x00 = [0.5]
x0 = torch.tensor(x00, dtype=torch.float32)
x0np = np.array(x00)
tn = 5.
ntime = 10000
ntrain = 5000
npaths = 1 # number of sample paths in data ensemble
npaths_fit = 5 # number of sample paths to generated under fitted model
seed = 17
lr = 0.001
weight_decay = 0.01 # Weight decay improves [32, 16] hidden dim fit by a lot!
epochs = 10000
hidden_dim = [2, 2]
num_layers = 2
noise_dim = 1
act = "Tanh"
printfreq = 1000
state_dim = x0.size()[0]
h = tn / ntime
torch.manual_seed(seed)
# Ground truth coefficients: One dimensional example: Mean-Reverting Process
def mu(t, x):
return 1.1 * (0.9 - x)
def sigma(t, x):
return 0.15 * np.sqrt(x)
# Generating ensemble data
sde = SDE(mu, sigma)
ensemble = sde.sample_ensemble(x0np, tn, ntime, npaths, noise_dim=noise_dim)
ensemble = torch.tensor(ensemble, dtype=torch.float32)
training_ensemble = torch.zeros((npaths, ntrain, state_dim))
test_ensemble = torch.zeros((npaths, ntime - ntrain + 1, state_dim))
for j in range(npaths):
training_ensemble[j, :, :] = ensemble[j, :ntrain, :]
test_ensemble[j, :, :] = ensemble[j, ntrain:, :]
# Neural SDE model to fit to ensemble data
nsde = NeuralSDE(state_dim, hidden_dim, num_layers, act, noise_dim)
nsde.fit(training_ensemble, lr, epochs, printfreq, h, weight_decay)
print("NLL on Test Ensemble = " + str(nsde.loss(test_ensemble, h)))
# Throw to an SDE object for convenient simulations of ensembles
sde_fit = SDE(nsde.mu_fit, nsde.sigma_fit)
ensemble_fit = sde_fit.sample_ensemble(x0np, tn, ntime, npaths=npaths_fit)
ensemble = ensemble.detach().numpy()
# Plot ensembles
fig = plt.figure()
t = np.linspace(0, tn, ntime + 1)
for i in range(npaths):
if state_dim == 2:
plt.plot(ensemble[i, :, 0], ensemble[i, :, 1], c="black", alpha=0.5)
elif state_dim == 1:
plt.plot(t, ensemble[i, :, 0], c="black", alpha=0.5)
for i in range(npaths_fit):
if state_dim == 2:
plt.plot(ensemble_fit[i, :, 0], ensemble_fit[i, :, 1], c="blue", alpha=0.5)
elif state_dim == 1:
plt.plot(t, ensemble_fit[i, :, 0], c="blue", alpha=0.5)
# Creating custom legend entries
true_line = plt.Line2D([], [], color='black', label='True')
model_line = plt.Line2D([], [], color='blue', label='Model')
# Adding the legend to the plot
plt.legend(handles=[true_line, model_line])
plt.show()