R code to reproduce the transcriptomic analysis of the Dias/Koup project

Fig. 6A-B: R code [MD]
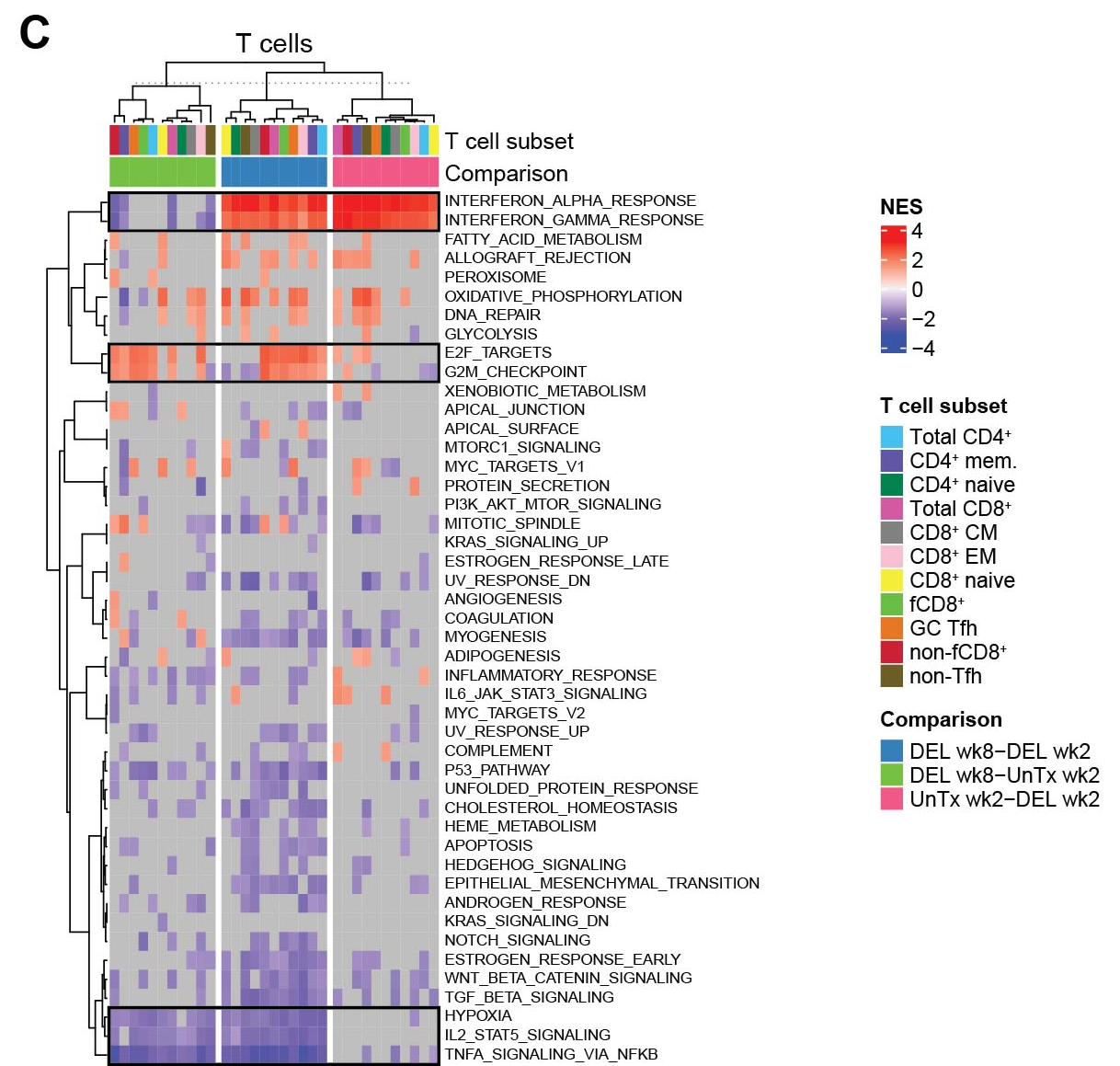 Fig. 6C: R code [MD]
Fig. 6C: R code [MD]
code:
input:
- non-normalized (raw) gene counts: [CSV]
- Rhesus genome annotation (see data release)
- alignment statistics: [TSV]
- sample annotation: [XLSX]
- viral load and bnAb concentrations: [XLSX]
output:
- raw SeqExpressionSet: [RDA]
- normalized SeqExpressionSet: [RDA]
- baseline-substracted SeqExpressionSet: [RDA]
- DGEGLM list: [RDA]
- GSEA output (see data release)
code:
- preprocessing: [MD]
input:
- non-normalized (raw) gene counts: [CSV]
- Rhesus genome annotation (see data release)
- alignment statistics: [TSV]
- sample annotation: [XLSX], [TSV]
output: