A toolbox for biosignal processing written in Python.
🌀 New module for signal quality assessment 🌀
With the biosppy.quality module you can now evaluate the quality of your signals!
So far, the EDA and ECG quality are available, but more could be added soon.
🫀 New module for heart rate variability (biosppy.signals.hrv)
🎊 New module for feature extraction (biosppy.features)
The toolbox bundles together various signal processing and pattern recognition methods geared towards the analysis of biosignals.
Highlights:
- Support for various biosignals: ECG, EDA, EEG, EMG, PCG, PPG, Respiration, HRV
- Signal analysis primitives: filtering, frequency analysis
- Feature extraction: time, frequency, and non-linear domain
- Signal quality assessment
- Signal synthesizers
- Clustering
- Biometrics
Documentation can be found at: https://biosppy.readthedocs.org/
Installation can be easily done with pip:
$ pip install biosppyAlternatively, you can install the latest version from the GitHub repository:
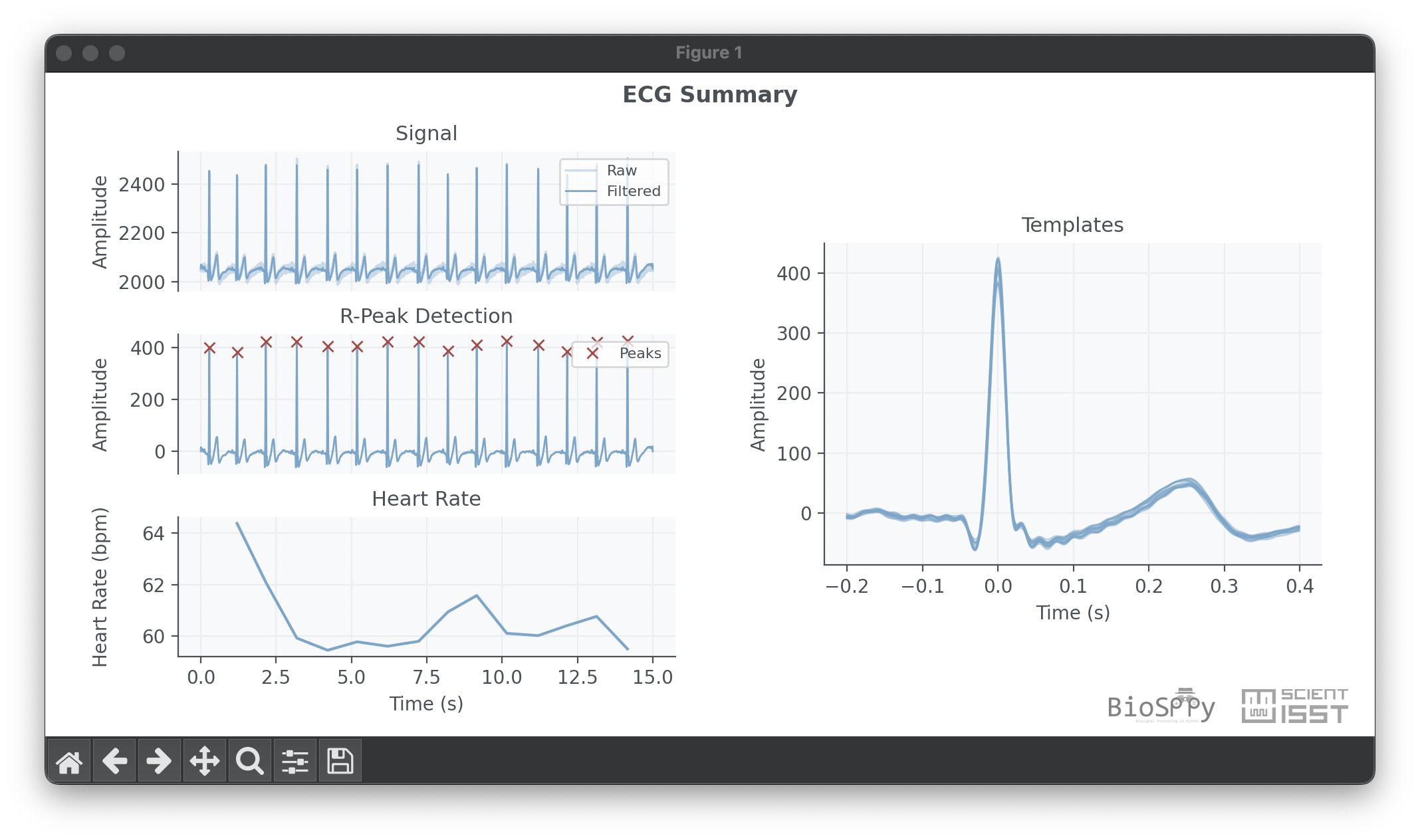
$ pip install git+https://github.com/scientisst/BioSPPy.gitThe code below loads an ECG signal from the examples folder, filters it,
performs R-peak detection, and computes the instantaneous heart rate.
from biosppy import storage
from biosppy.signals import ecg
# load raw ECG signal
signal, mdata = storage.load_txt('./examples/ecg.txt')
# process it and plot
out = ecg.ecg(signal=signal, sampling_rate=1000., show=True)This should produce a plot similar to the one below.
- bidict
- h5py
- matplotlib
- numpy
- scikit-learn
- scipy
- shortuuid
- six
- joblib
Please use the following if you need to cite BioSPPy:
P. Bota, R. Silva, C. Carreiras, A. Fred, and H. P. da Silva, "BioSPPy: A Python toolbox for physiological signal processing," SoftwareX, vol. 26, pp. 101712, 2024, doi: 10.1016/j.softx.2024.101712.
@article{biosppy,
title = {BioSPPy: A Python toolbox for physiological signal processing},
author = {Patrícia Bota and Rafael Silva and Carlos Carreiras and Ana Fred and Hugo Plácido {da Silva}},
journal = {SoftwareX},
volume = {26},
pages = {101712},
year = {2024},
issn = {2352-7110},
doi = {https://doi.org/10.1016/j.softx.2024.101712},
url = {https://www.sciencedirect.com/science/article/pii/S2352711024000839},
}However, if you want to cite a specific version of BioSPPy, you can use Zenodo's DOI:
BioSPPy is released under the BSD 3-clause license. See LICENSE for more details.
This program is distributed in the hope it will be useful and provided to you "as is", but WITHOUT ANY WARRANTY, without even the implied warranty of MERCHANTABILITY or FITNESS FOR A PARTICULAR PURPOSE. This program is NOT intended for medical diagnosis. We expressly disclaim any liability whatsoever for any direct, indirect, consequential, incidental or special damages, including, without limitation, lost revenues, lost profits, losses resulting from business interruption or loss of data, regardless of the form of action or legal theory under which the liability may be asserted, even if advised of the possibility of such damages.