tflow::use_tflow()
✔ Setting active project to '/home/rmflight/Projects/personal/example_targets_workflow'
✔ Creating 'R/'
✔ Writing 'packages.R'
✔ Writing '_targets.R'
✔ Writing '.env'Put the data files in there.
dir("data")
[1] "sample_measurements.csv" "sample_metadata.csv"> tar_make()
• start target measurement_file
• built target measurement_file
• start target metadata_file
• built target metadata_file
• end pipeline: 0.06 secondstar_make()
✔ skip target measurement_file
✔ skip target metadata_file
• start target lipid_measurements
Rows: 1012 Columns: 16
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (4): class, name, group_units, feature_id
dbl (12): WT_1, WT_2, WT_3, WT_4, WT_5, WT_6, KO_1, KO_2, KO_3, KO_4, KO_5, ...
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
Rows: 12 Columns: 18
── Column specification ────────────────────────────────────────────────────────
Delimiter: ","
chr (10): parent_sample_name, assay, cell_line, client_matrix, client_sample...
dbl (8): client_identifier, client_sample_number, group_number, sample_amou...
• built target lipid_measurements
• start target lipid_metadata
\
ℹ Use `spec()` to retrieve the full column specification for this data.
ℹ Specify the column types or set `show_col_types = FALSE` to quiet this message.
• built target lipid_metadata
• end pipeline: 0.456 seconds> tar_make()
✔ skip target measurement_file
✔ skip target metadata_file
✔ skip target lipid_measurements
✔ skip target lipid_metadata
• start target exploration
• built target exploration
• end pipeline: 2.606 second> tar_make()
✔ skip target measurement_file
✔ skip target metadata_file
✔ skip target lipid_measurements
✔ skip target lipid_metadata
• start target lipid_normalized
• built target lipid_normalized
✔ skip target exploration
• start target lipid_imputed
• built target lipid_imputed
• start target lipids_differential
• built target lipids_differential
• end pipeline: 0.118 seconds> tar_make()
✔ skip target measurement_file
✔ skip target metadata_file
• start target differential_report
• built target differential_report
✔ skip target lipid_measurements
✔ skip target lipid_metadata
✔ skip target lipid_normalized
✔ skip target exploration
✔ skip target lipid_imputed
✔ skip target lipids_differential
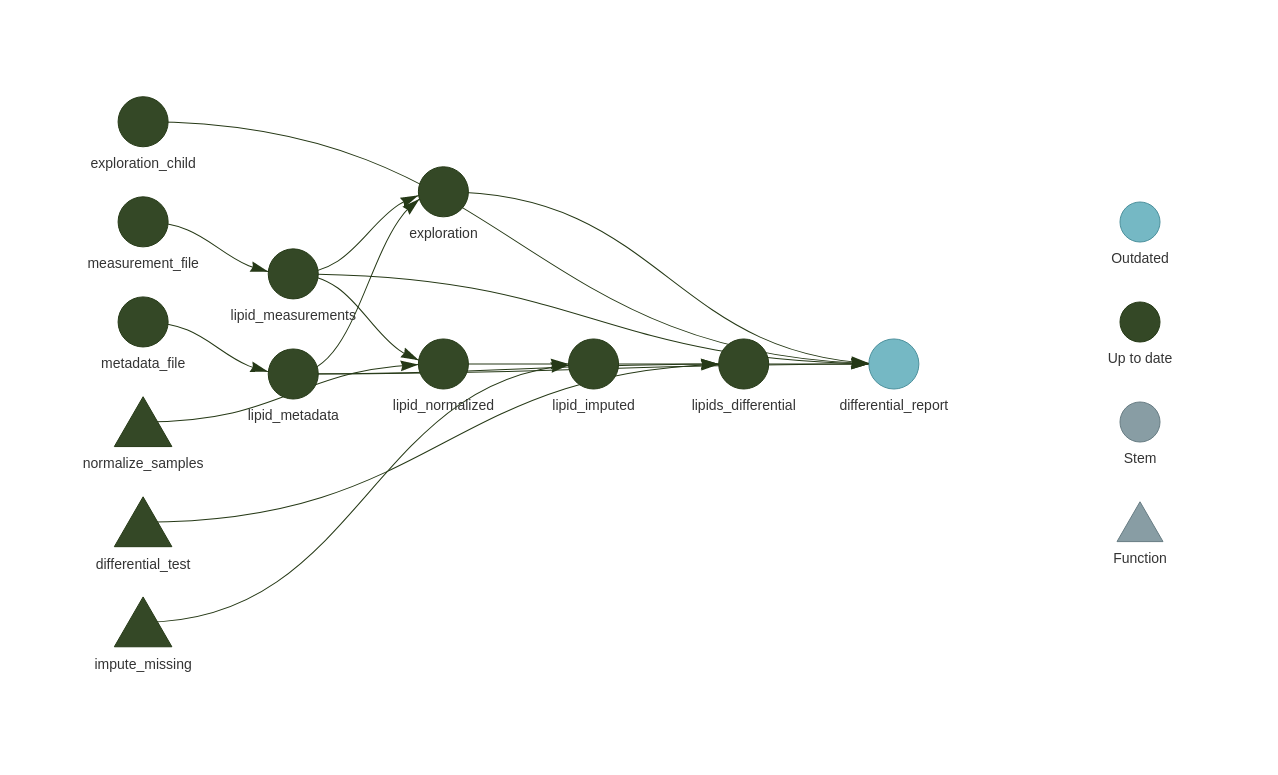
• end pipeline: 0.63 secondsIf you wanted to always make sure that the EDA full report got rendered first, then you can add it to the tar_load call at the top of the differential_report.Rmd.
tar_load(c(exploration,
exploration_child,
lipids_differential))This makes it part of the dependency graph for the differential_report.