Harmony is a tool used to correct batch effects in single-cell RNA seq datasets.
To use the Harmony module, you will need to have put your scRNA-seq data through the Seurat pipeline available on GenePattern (Seurat.QC --> Seurat.Preprocessing). The user must input more than two Seurat objects contained in RDS files. Once Harmony has been completed, the module will output four files:
Harmonized Data - An RDS file containing a Seurat object with the Harmony-processed data. The Harmony-adjusted principal components can be found in the "harmony" column under the "reduction" slot in the Seurat object. The name of this file is specified by the "Output Name" parameter.
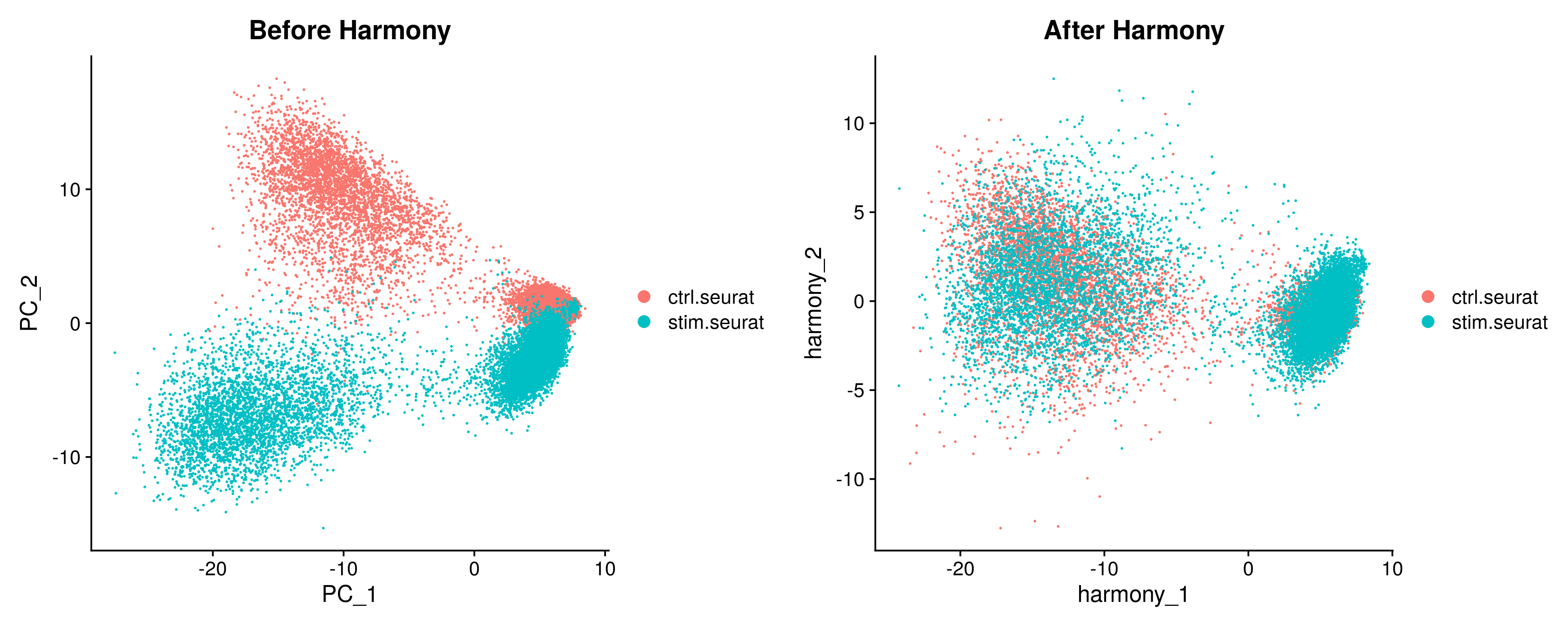
Before Harmony Plot - A PNG file showing a scatterplot of the dimensionality-reduced data before Harmony. The method of dimensionality-reduction shown can be specified by the "reduction" parameter.
After Harmony Plot - A PNG file showing a scatterplot of the data post-Harmony.
Side To Side Plot - A PNG file showing the Before Harmony Plot and After Harmony Plot side by side for debugging purposes.
Here are the basic parameters you will need in order to run the Harmony module.
-
Input RDS Files (required) - The list of datasets you wish to analyze with Harmony. Each dataset must consist of one group of data, and must be a Seurat object in an .rds file.
-
Output Name (required) - The prefix that you would like to use to name your output files. One file will contain your Harmony-processed data, and another file will display the scatterplot made for the data.
-
Cell Types (optional) - The names of the datasets you would like to run Harmony on. The list of names should be as long as the list of files. By default, the names of the cell types will be designated as the names of the files.
-
Group Name (optional) - The name of the metadata column you would like to group by during visualization. If no group name is specified, then Harmony will group by dataset by default.
-
Colors (optional) - The colors you want to use to label your groups during visualization. If no colors are specified, then they will be automatically assigned. Refer to this link to see the list of color options to choose from: https://www.datanovia.com/en/blog/awesome-list-of-657-r-color-names/
These parameters are for more advanced use. They are all optional.
-
reduction - Name of dimension reduction to use. Default: 'pca'
-
dims use - Which PCA dimensions to use for Harmony. By default, use all
-
theta - Diversity clustering penalty parameter. Specify for each variable in group.by.vars. theta=0 does not encourage any diversity. Larger values of theta result in more diverse clusters. Default: 2
-
lambda - Ridge regression penalty parameter. Specify for each variable in group.by.vars. Lambda must be strictly positive. Smaller values result in more aggressive correction. Default: 1
-
sigma - Width of soft kmeans clusters. Sigma scales the distance from a cell to cluster centroids. Larger values of sigma result in cells assigned to more clusters. Smaller values of sigma make soft kmeans cluster approach hard clustering. Default: 0.1
-
nclust - Number of clusters in model. nclust=1 equivalent to simple linear regression.
-
tau - Protection against overclustering small datasets with large ones. tau is the expected number of cells per cluster. Default: 0
-
block size - What proportion of cells to update during clustering. Between 0 to 1. Larger values may be faster but less accurate. Default: 0.05
-
max iter harmony - Maximum number of iterations that Harmony will run. Default: 10
-
max iter cluster - Maximum number of rounds to run clustering at each round of Harmony. Default: 20
-
stop early cluster - Whether or not to stop clustering early. If TRUE, then the convergence tolerance is specified by the epsilon cluster parameter. Default: TRUE
-
epsilon cluster - Convergence tolerance for clustering round of Harmony. Default: 0.00005
-
stop early harmony - Whether or not to stop harmony early. If TRUE, then the convergence tolerance is specified by the epsilon harmony parameter. Default: TRUE
-
epsilon harmony - Convergence tolerance for Harmony. Default: 0.0004
-
plot_convergence - Whether to print the convergence plot of the clustering objective function. TRUE to plot, FALSE to suppress. This can be useful for debugging. Default: FALSE
-
verbose - Whether to print progress messages. TRUE to print, FALSE to suppress. Default: TRUE
-
reference_values - Defines reference dataset(s). Cells that have batch variables values matching reference_values will not be moved
-
reduction save - Keyword to save Harmony reduction. Useful if you want to try Harmony with multiple parameters and save them as e.g. 'harmony_theta0', 'harmony_theta1', 'harmony_theta2'. Default: harmony
-
assay use - Which assay to run PCA on if no PCA present?
-
project dim - Project dimension reduction loadings. Default: TRUE
- Harmony was developed by the Raychaudhuri Lab.
- Original Harmony GitHub repo: https://github.com/immunogenomics/harmony
- Harmony paper: https://pubmed.ncbi.nlm.nih.gov/31740819/
- Docker image used: https://hub.docker.com/r/jzl010/harmony
Ilya Korsunsky, Nghia Millard, Jean Fan, Kamil Slowikowski, Fan Zhang, Kevin Wei, Yuriy Baglaenko, Michael Brenner, Po-Ru Loh, Soumya Raychaudhuri
Justin Lee: [email protected]