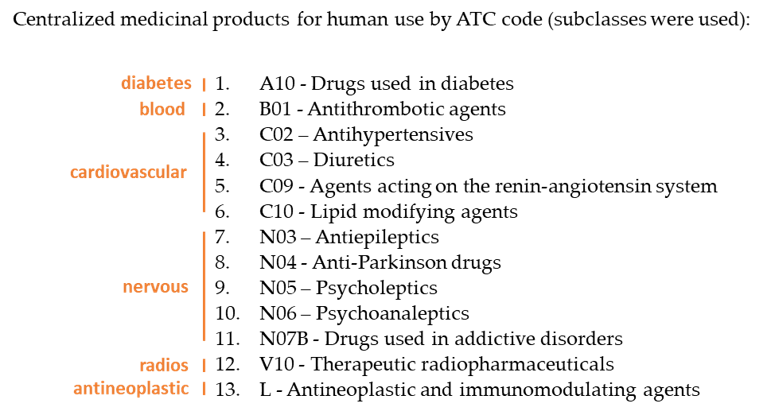Predicting patient mortality within 180 days after their last hospital visit based on Electronic Health Record data
Team: moore
Panagiotis Togias, Maria-Theodora Pandi
An XGBOOST [2] implementation for patient mortality prediction in a 180-day window since their last hospital visit using EHR data and comorbidities scores.
The objective of this challenge was to develop a method that uses curated EHR data to predict patient mortality, within 6 months from the last recorded visit in a healthcare facility. The data follow the OMOP v5.0 format, which is used for the integration and standardization of clinical information, and consist of 8 tables of demographic and clinical features, plus one more table that contains the death records for the training dataset. When approaching this challenge, we choose to train an XGBOOST model, as it is considered as a top performing algorithm in a variety of applications, while our main efforts focused on data preprocessing. In the original format of the data, multiple entries across different tables may exist for each patient, corresponding to different visits, conditions, medications he/she has received etc. Our main purpose was to summarize them into a single record per patient, while maintaining as much information as possible, and then to convert the categorical variables into numeric, so they can be used to train the XGBOOST model. Following the process described in the Methods section, we focused on the concept_id column of each table of interest and replaced individual values with broader categories, which correspond to features that indicate an overall impaired health status. Subsequently, for every patient, we calculated the appearances of each variable. We applied this approach in the 3rd round of the Challenge and observed an extreme improvement of our model’s predictive power, based on AUROC. Replacing our first quite arbitrary definition of these categories, during the extension period, and exploiting information from the Athena database, lead to further significant improvements in both AUROC and AUPR scores.
| Approach | Challenge period | AUROC | AUPR |
|---|---|---|---|
| Feature engineering using the Athena Database | Extension of phase 3 | 0.9372 | 0.1552 |
| Feature engineering using information coming from “fast lane” dataset | Phase 3 | 0.9134 | 0.0865 |
| Sums and counts of non-NA clinical features of interest | Phase 2 | 0.51365 | 0.007252 |
Note: Although a big number of submission models returned high AUROC scores (> 0.9), we are a bit skeptical about the correct prediction of both classes (mortality/no mortality-mortality after 180 days) because of the significant class imbalance that is present in the dataset. This left us wondering whether these high scores are in fact product of the correct prediction of the larger proportion of True Negatives (no mortality-mortality after 180 days) over the much less True Positives (mortality). Spaces such as the PR tend to be robust to these kind of issues, as they account only for the True Positive class, and give a more clear view regarding algorithm prediction performance.
In order to transform multiple individual patient records into a single entry for each person_id, we relied on features expected to have a negative impact in health and counted each recorded instance per patient. In addition, some demographics were also considered such as age, gender and race, while ethnicity was not included in the final model due to its’ high cardinality. In addition, we focused on Observations, to extract information related to the ability of a patient to pay his/her medical expenses. All concept vocabularies were downloaded though the OHDSI Athena database and an extensive search was performed in it in order to create the previously mentioned features considering the respective vocabulary for each table.
Bellow we present each variable (46 in total) and its respective table of origin as well as the method used for engineering it:
| Variable Name | Variable Description | Origin Table | Engineering Method |
|---|---|---|---|
| cancer | Any non-metastatic form of cancer | Condition_occ | Matching String Search |
| cancermetastatic | Metastatic cancer | Condition_occ | Matching String Search |
| diabetes | Diabetes Mellitus | Condition_occ | Matching String Search |
| alzheimersdementia | Alzheimer’s and Dementia-related Diseases | Condition_occ | Matching String Search |
| cerebrovascular | Cerebrovascular-related conditions | Condition_occ | Matching String Search |
| heartfailure | Conditions recorded as “Heart Failure” | Condition_occ | Matching String Search |
| coronarysyndrome | Coronary syndrome | Condition_occ | Matching String Search |
| myocardialinfarction | Myocardial infarction | Condition_occ | Matching String Search |
| immunodeficiencies | Immunodeficiency-related conditions | Condition_occ | Matching String Search |
| respiratoryfailure | Respiratory failure | Condition_occ | Matching String Search |
| endstageconditions | End stage flagged conditions | Condition_occ | Matching String Search |
| adverseeffects | Conditions mentioned as “Adverse effects” or “Adverse reactions” | Condition_occ | Matching String Search |
| copd | Chronic Obstructive Pulmonary Disease (Emphysema, Chronic Bronchitis) | Condition_occ | Matching String Search |
| hypertension | Hypertension-related conditions | Condition_occ | Matching String Search |
| drugabuse | Drug abuse, as defined by Elixhauser | Condition_occ | Elixhauser comorbidities |
| weightloss | Weight loss, as defined by Elixhauser | Condition_occ | Elixhauser comorbidities |
| coagulopathy | Coagulopathy, as defined by Elixhauser | Condition_occ | Elixhauser comorbidities |
| liverdisease | Liver disease, as defined by Elixhauser | Condition_occ | Elixhauser comorbidities |
| renalfailure | Renal failure, as defined by Elixhauser | Condition_occ | Elixhauser comorbidities |
| neurological | Neurological disorders, as defined by Elixhauser | Condition_occ | Elixhauser comorbidities |
| pulmonarycirculation | Pulmonary circulation disorders | Condition_occ | Elixhauser comorbidities |
| paralysis | Paralysis, as defined by Elixhauser | Condition_occ | Elixhauser comorbidities |
| fluidelectrolyte | Fluid and electrolyte disorders, as defined by Elixhauser | Condition_occ | Elixhauser comorbidities |
| psychosesdepression | Combined Physchoses and Depresssion, as defined by Elixhauser | Condition_occ | Elixhauser comorbidities |
| transfusions | All transfusion-related procedures | Procedure_occ | Matching String Search |
| heartbypass | All heart bypass-related procedures | Procedure_occ | Matching String Search |
| cardiacpacemaker | All cardiacpacemaker-related procedures | Procedure_occ | Matching String Search |
| transplants | All transplant-related procedures | Procedure_occ | Matching String Search |
| hemodialysis | All hemodialysis-related procedures | Procedure_occ | Matching String Search |
| radios_d | Active compounds used as radiotherapeutics or -diagnostics | Drug_exposure | ATC Ingredient Category |
| antineoplastic_d | Antineoplastic compounds | Drug_exposure | ATC Ingredient Category |
| blood_d | Active compounds used for Blood-related conditions | Drug_exposure | ATC Ingredient Category |
| cardiovascular_d | Active compounds used for Cardiovascular conditions | Drug_exposure | ATC Ingredient Category |
| diabetes_d | Active compounds used for Diabetes Mellitus | Drug_exposure | ATC Ingredient Category |
| nervous_d | Active compounds used for neurological conditions | Drug_exposure | ATC Ingredient Category |
| obese | Observations indicating obesity (based on BMI – boolean variable) | Observation_occ | Matching String Search |
| deathobservations | Observations that included any information about someone’s death (e.g. time of death, etc. – boolean variable) | Observation_occ | Matching String Search |
| ecscore | A variation of the Elixhauser comorbidity score | Elixhauser comorbidity score | |
| unabletopay | Boolean variable indicating whether a patient has any record of not being able to pay medical bills / is not insuranced / has bad financial status | Observation_occ | Matching String Search |
Variables regarding summations of each table per patient
| Variable Name | Variable Description |
|---|---|
| visits_sum | Sum of recorded visits per patient |
| observation_days | Recorded observation days per patient |
| procedures_sum | Sum of recorded procedures per patient |
| measurements_sum | Sum of recorded measurement instances per patient |
| conditions_sum | Sum of recorded conditions per patient |
| drugs_used_sum | Sum of recorded drugs used per patient |
| drug_exposure_days | Sum of recorded days of exposure to drugs per patient |
A string search in Athena database, containing terms of interest, in order to extract the respective concept ids and engineer each variable that falls under this category. For this procedure to obtain only useful information and exclude noise from entries implicating occurrences with no value but including the string of interest, restrictions were implemented where possible.
For example, this is the case of constructing the “cancermetastatic” variable by searching all SNOMED descriptions in Athena database:
domain[domain_id == "Condition" &
vocabulary_id == "SNOMED" &
((concept_name %ilike% "malignant" &
(concept_name %ilike% "tumor" | concept_name %ilike% "neoplasm")) |
(concept_name %ilike% "sarcoma" | concept_name %ilike% "carcinoma" | concept_name %ilike% "lymphoma" | concept_name %ilike% "leukemia" | concept_name %ilike% "Hodgkin") |
(concept_name %ilike% "cancer")|
(concept_name %ilike% "figo" & concept_name %ilike% "iva|ivb")) &
(concept_name %ilike% "secondary" | concept_name %ilike% "metasta" | concept_name %ilike% "stage 4|stage iv") &
!(concept_name %ilike% "history" | concept_name %ilike% "remission") &
!(concept_name %ilike% "fear" | concept_name %ilike% "phobia" | concept_name %ilike% "carrier" | concept_name %ilike% "hereditary" | concept_name %ilike% "cancer screening")]
Comorbidities proposed by Elixhauser et. al. [4] were considered here, because of their recent publication date in contrast with the Charlson comorbidity score [3]. Both scores were introduced with the ICD10 vocabulary, so we transformed them to SNOMED for them to be concise with the rest variables regarding conditions. Weights for the Elixhauser comorbidity score were calculated based on a variation of the AHRQ Algorithm [5] that we applied. In this specific scenario, we emphasized on conditions that are generally known to have a negative impact on a person’s health based on today’s medical standards and assigned them a weight based on the process described by AHRQ Algorithm. The weights for each person are then summed for a single score to be produced (0 the lowest score and 90 the highest score that a person can have). We chose to leave some scores as they were originally proposed in literature.
More Specifically:
| Variable Name | Weight |
|---|---|
| heartfailure | 9 |
| pulmonarycirculation | 6 |
| paralysis | 5 |
| neurological | 5 |
| copd | 3 |
| renalfailure | 6 |
| liverdisease | 4 |
| cancermetastatic | 14 |
| cancer | 7 |
| coagulopathy | 11 |
| weightloss | 9 |
| fluidelectrolyte | 11 |
Except from conditions, procedures and some other relevant observations, a patient’s drug exposure must be recorded in a way that is well understood and categorized. For this reason, we went with the ATC (Anatomical Therapeutic Chemical) classification system. Quoting the respective Wikipedia post:[6] “ATC is a drug classification system that classifies the active ingredients of drugs according to the organ or system on which they act and their therapeutic, pharmacological and chemical properties. It is controlled by the World Health Organization Collaborating Centre for Drug Statistics Methodology (WHOCC) and was first published in 1976”. Once we had a list of all 14 top-level classes, a further selection took place in order to retain those that were considered to have the most negative impact in a person’s health or were associated with high risk conditions.
The Drug_exposure data follow the RxNorm vocabulary, which is formed using the active ingredient of a drug, its’ strength and dose form. Again, we performed a string search of the ingredients (following the categorization of ATC) in the RxNorm vocabulary.
For example, the search regarding the “radios” variable was conducted like this:
domain[domain_id == "Drug" &
vocabulary_id == "RxNorm" &
concept_class_id == "Ingredient" &
concept_name %ilike% "samarium|tiuxetan|radium|lutetium"]
Before moving into the training phase, we included a few more steps in our preprocessing pipeline. Here is a brief list of these matters:
- “Obese”, “deathobservations” and gender variables were converted to Boolean (presence/absence)
- Boolean variable “unabletopay” was engineered through four other previously created variables indicating the recorded financial and insurance status of each patient (“finstatus_0”, “finstatus_1”, “insurance_0”, “insurance_1”). 1 suggesting that a patient has a record of not being able to pay medical expenses and 0 when a patient has a record of being able to pay
- All NA values (except the ones in columns “age”, “gender_concept_id”, “race_concept_id” and those used for feature engineering the “unabletopay” variable) were replaced with zeros
- For each person’s race, a scale form 1-6, allowing for NAs was created to represent the 6 major racial groups based on the Race and Ethnicity Code Set (USBC)
- Death indication was removed and changed back to “alive” if 180 days have passed since their last visit to a health care unit after the recorded death date
- Variables with zero variance were removed
The same preprocessing steps were applied in both training and inferring sets.
Two XGBOOST models were trained in our last submission in round 3 extention with the best one achieving an AUROC score = 0.9372. In order to test one more scenario regarding model parameter tuning, we decided, in our final submission in the evaluation phase, to add a third one and see how that goes later on. For the selection of optimal nrounds, we separated again the existing training set to additional train / eval sets (93% / 7% respectively) based on the distribution of the target variable and used the new evaluation set for the selection of an optimal number of nrounds through XGBOOST's watchlist feature.
| 1st model (3ext) | 2nd model (3ext) | 3rd model (eval) |
|---|---|---|
| Evaluation Metric | auc | auc |
| nrounds | 1000 | 1000 |
| max_depth | 4 | 4 |
| scale_pos_weight | sqrt(sum of alive / sum of dead) | sum of alive / sum of dead |
| Learning Rate | 0.01 | 0.1 |
| tree_method | exact | hist |
| objective | binary:logistic | binary:logistic |
| min_child_weight | 0 | 0 |
| **early_stopping_rounds ** | 100 | 100 |
Notes on parameter tuning : We tested _aucpr _ as the evaluation metric on other submissions but surprisingly it ended up giving slightly worse results. _logloss _is also a robust evaluation metric when it comes to class imbalance but was not included in this work.
As mentioned earlier, the problem of class imbalance was an issue in this challenge. In order to have a better understating of how models behave on balanced EHR data, downsampling the majority class should give a more clear feedback on the correct prediction of both classes. Nevertheless, this is a great opportunity to create solid ground for better future work.
Our approach emphasized more on a feature engineering perspective of patient morality prediction. Having a look at the AUROC and AUPR scores of other participants higher on the leaderboard, made us consider whether we missed something when engineering variables. Maybe an even more extensive search for variables may be appropriate in future analyses or other features that were not included in this challenge are the key for better results. Another aspect that should be investigated is the new version (v6.0) of OMOP Common Data Model that offers a few updates on tables and variables.
Some final remarks:
- Many different OMOP ids refer to the same concept making the exploitation of EHR datasets for predictive and machine learning purposes difficult
- Predictions are made based on overall health / not a disease-specific application
- Less categories of the concepts (e.g. in ICD-10 vocabulary) seem to be more informative and easier to work with
Future directions: - Need to explore better the effects of comorbidity scores (Elixhauser’s, Charlson’s etc) in such approaches
- Expert based definition (or relying more on published literature) of the new variables that we consider to be informative in terms of mortality prediction and the concepts they include
- For cancer, the explicit use of the International Classification of Diseases for Oncology (ICD-O-3) may be more appropriate
- Apply dimensionality reduction to highly correlated variables
[1] EHR DREAM Challenge: Patient Mortality (syn18405991)
[2] Chen T & Guestrin C. XGBoost: A Scalable Tree Boosting System. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (KDD ’16). Association for Computing Machinery, New York, NY, USA. 2016; 785–794. DOI:10.1145/2939672.2939785
[3] Charlson ME, Pompei P, Ales KL, et al. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. Journal of Chronic Diseases 1987; 40:373-383. DOI:10.1016/0021-9681(87)90171-8
[4] Elixhauser A, Steiner C, Harris DR and Coffey RM. Comorbidity measures for use with administrative data. Medical Care 1998; 36(1):8-27. DOI:10.1097/00005650-199801000-00004
[5] Moore BJ, White S, Washington R, Coenen N, and Elixhauser A. Identifying increased risk of readmission and in-hospital mortality using hospital administrative data: the AHRQ Elixhauser comorbidity index. Medical Care 2017; 55(7):698-705. DOI:10.1097/MLR.0000000000000735
[6] [Online Source] Wikipedia, Anatomical Therapeutic Chemical Classification System, https://en.wikipedia.org/wiki/Anatomical_Therapeutic_Chemical_Classification_System, Accessed on: Jan 7th, 2020