USUM uses USEARCH and UMAP (or t-SNE) to plot DNA 🧬 and protein 🧶 sequence similarity embeddings.
-
Install
USEARCHdependency manually: https://drive5.com/usearch/download.html
(consider supporting the author by buying the 64bit license) -
Install
usumusing PIP:
pip install usumUse usum to plot input protein or DNA sequences in FASTA format.
Show all available options using usum --help
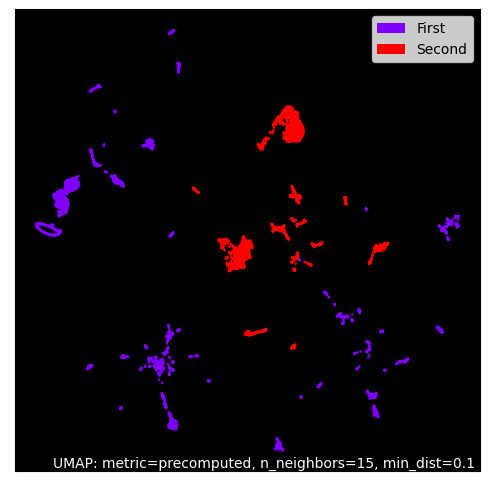
usum example.fa --maxdist 0.2 --termdist 0.3 --output exampleusum first.fa second.fa --labels First Second --maxdist 0.2 --termdist 0.3 --output exampleThis will produce a PNG plot:
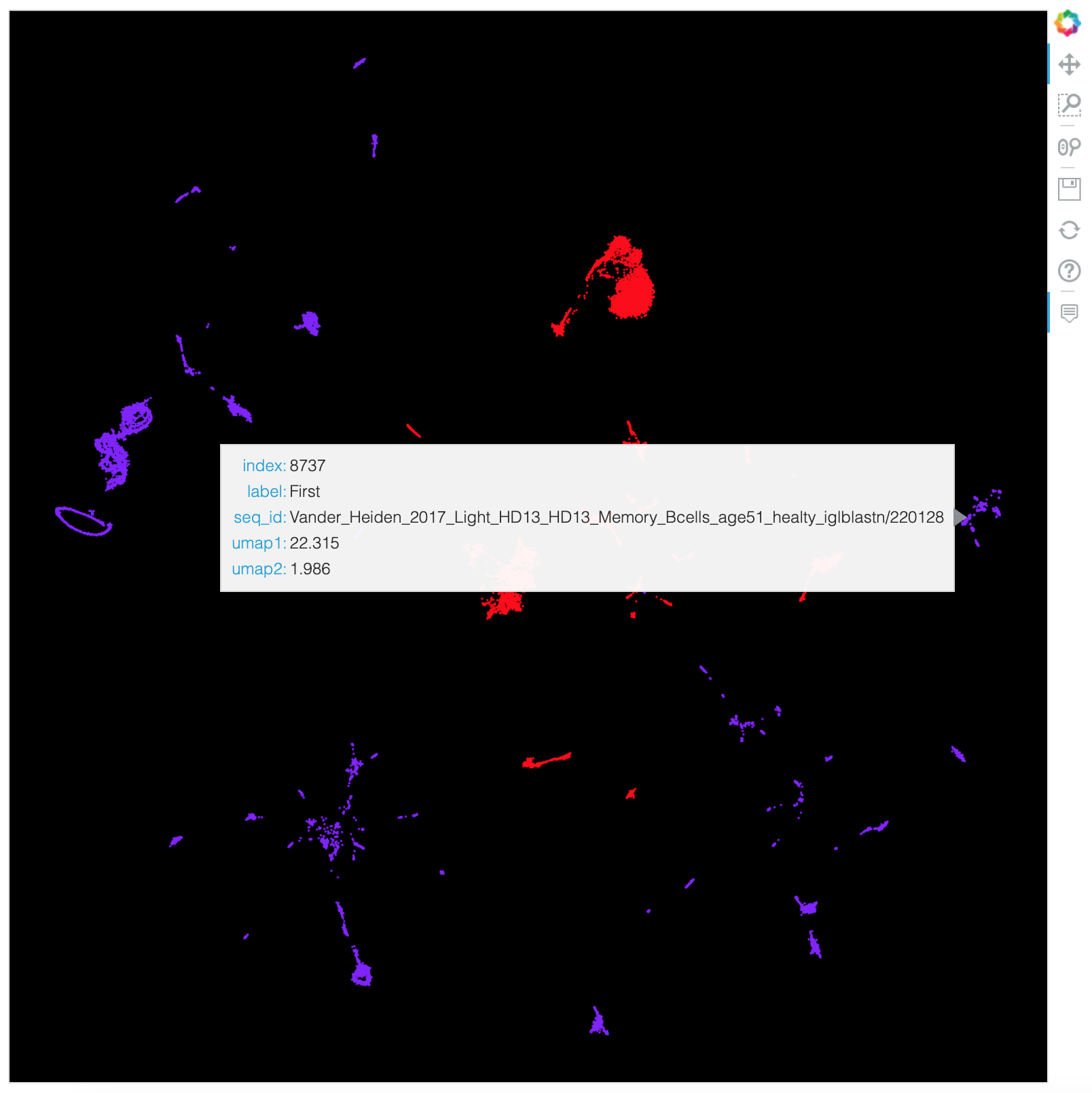
An interactive Bokeh HTML plot is also created:
You can also produce a t-SNE plot using the --tsne flag.
usum first.fa second.fa --labels First Second --maxdist 0.2 --termdist 0.3 --tsne --output exampleThis will produce a PNG plot:
You can use --limit to extract and plot a random subset of the input sequences.
# Plot 10k sequences from each input file
usum first.fa second.fa --labels First Second --limit 10000 --maxdist 0.2 --termdist 0.3 --output exampleYou can control randomness and reproducibility using the --seed option.
See usum --help for all plotting options.
See UMAP API Guide for more info about the UMAP options.
- Use
--limitto plot a random subset of records - Use
--widthand--heightto control plot size in pixels - Use
--resumeto reuse previous distance matrix from the output folder - Use
--tsneto produce a t-SNE embedding instead of UMAP (you can use this with--resume) - Use
--umap-spreadto control how close together the embedded points are in the UMAP embedding - Use
--umap-min-distto control minimum distance between points in UMAP embedding - Use
--neighborsto control number of neighbors in UMAP graph
When changing just the plot options, you can use --resume to reuse previous results from the output folder.
Warning This will reuse the previous distance matrix, so changes to limits or USEARCH args won't take effect.
# Reuse result from umap output directory
usum --resume --output example --width 600 --height 600 --theme firefrom usum import usum
# Show help
help(usum)
# Run USUM
usum(inputs=['input.fa'], output='usum', maxdist=0.2, termdist=0.3)- A sparse distance matrix is calculated using USEARCH calc_distmx command.
- The distances are based on % identity, so the method is agnostic to sequence type (DNA or protein)
- The distance matrix is embedded as a
precomputedmetric using UMAP - The embedding is plotted using umap.plot.