This work is part of the Software Project: "Language, Action and Perception" at Saarland Universitz, WS 2020-2021.
This repository contains our implementation and a summary of our research findings.
- Directory Organization
- Project Overview
- MMBT Model
- Dataset
- Requirements
- Instructions
- Notebooks
- Preprocess
- Experiments
- Results
- References
This project repository is organized as follows:
- Pre-Trained Multi-Modal Text and Image Classifier in Sparse Data Application: the parent project directory
- this README.md file
- data/: contains data files subdirectories and data preparation scripts
- json/
- csv/
- image_labels_csv/
- models/
- NLCXR_front_png/
- preparations.py
- MMBT/: contains MMBT model src codes and related utility functions
- image.py
- mmbt.py
- mmbt_config.py
- mmbt_utils.py
- runs/: saved Tensorboards for displaying models' performance
- integrated_gradients/:
- examples/
- figures/
- results/
- integrated_gradients.py
- main.py
- utils.py
- visualization.py
- run_mmbt.ipynb notebook
- run_bert_text_only.ipynb notebook
- run_mmbt_masked_text_eval.ipynb notebook
- image_submodel.ipynb notebook
- bertviz_attention.ipynb notebook
- false_preds.ipynb notebook
- baseline_experiments_results.ipynb notebook
- textBert_utils.py src codes and utility functions for the run_bert_text_only.ipynb notebook
Note: The NLCXR_front_png directory is NOT provided; please make this directory after cloning the repo and obtain the image files according to the instruction in the /data directory.
Note2: Previous run's outputs and checkpoints are omitted due to large file size. When new experiments are run, the notebooks will make a new directory in this parent directory. the runs/ directory will also be updated during each experiment (text-only and MMBT).
This repo presents our multimodal model, its submodules, and the experiments we performed to answer our research questions. Our main focus of the project is to try to answer the following questions:
- Can multimodal embeddings improve transfer learning performance in low-data classification tasks over unimodal embeddings?
- Related works show improvements in classification and similarity tasks when combining visual and text modalities. Does this effect carry over to the medical domain?
- How does the multimodal representation contribute to the improved performance?
- How do different approaches to combining modalities affect performance?
- Specifically, does having a more integrated fusion approach between the text and visual modalities perform better in downstream tasks than simply concatenating vectors representing the two modalities?
The experiments we ran on the model are as follows:
- Image-only classification
- Text-only classification
- Multimodal classification: text and image inputs
- Attention mechanism visualization
- Image-only classification with the multimodal model trained on text and image data
In addition, we also present the Integrated Gradient to visualize and extract explanations from the images.
-
We implemented our models in PyTorch and used Huggingface
BERT-base-uncasedmodel in all our BERT-based models. -
For training the MMBT model, we used the default settings for hyperparameters as in the original Huggingface Implementation
-
For training the text-only model, we used the default settings for hyperparameters as in Huggingace Example
-
We trained with the batch size of 32 and in some cases where memory is insufficient, 16, for the MMBT and text-only model.
-
We extended our training for the MMBT model to 10.0 epochs instead of the default = 3.0 in Huggingface. We noticed that MMBT model's validation starts to worsen (overfit) only after 6 epochs.
-
We trained the text only model to 4.0 epochs as recommended in the BERT paper by Devlin et al. (2019).
-
We followed the default hyparameter settings for DenseNet in fine-tuning the image-only model.
In our project, we are experimenting with the Supervised Multimodal BiTransformers for Classifying Images and Text (MMBT) presented by Kiela et al. (2020). This is a BERT-based model that can accommodate multi-modal inputs. The model aims to fuse the multiple modalities as the input data are introduced to the Transformers architecture so that the model's attention mechanism can be applied to the multimodal inputs. For more information regarding the model, please refer to the documentation in the MMBT directory.
For more information regarding the dataset utilized in the project and the preparation steps, please consult the information in the data directory.
Before proceeding to train and test the model, the frontal X-ray image files used in the experiments can be obtained via the link to this Shared Google Drive.
For access to all X-ray images, include non-frontal images, please refer to this Other Shared Google Drive.
The frontal images should be saved to the data/NLMCXR_png_frontal subdirectory inside the ./data/ directory.
The X-ray images from the other shared Google Drive should be saved to the data/NLMCXR_png subdirectory inside the ./data/ directory.
Please note that these 2 subdirectories are NOT included in this repo and will need to be created as part of the preparation steps to reproduce the experiments.
For reference, here's also a link to the original dataset of the Indiana University Chest X-ray.
- The project scripts were tested on Python 3.6 and 3.7. on Mac OS and Linux.
- We tested in Anaconda python virtual environment.
- Notebooks were tested on Google Colab and experiments were run on Google Colab when GPU is required.
- GPU is recommended to run the experiment. A single GPU is sufficient.
- the MMBT experiments will run out of standard CPU memory in Google Colab unless GPU is available.
- to run multiple experiments, it may be necessary to reset the runtime from time to time.
- Approximate runtime/experiment: 5-30 minutes.
-
Please create a virtual environment with the provided .yml file
LAP_environment.yaml
1.1 This.yamlfile was created on and for a conda environment on a Mac OSX-64 platform. If you are running a different system, this option may not work.
1.2 We have also made a less specific.yamlfile with only the necessary version requirements which is more likely to work on a non-OSX-64 platform.
1.3 Alternatively, you may also re-create an environment using conda or pip with on of the provided requirement files listed below; the less specific version should work across platform:
1.3.1requirements.txthas all the builds and version information and should work on an OSX-64 platform
1.3.2requirements_no_builds.txtonly has the version specifications for the modules and dependencies
1.3.3requirements_bare.txtonly has the list of modules (no dependencies) with no version information
1.4 see Alternative Instructions for replicating this project via Google Colab -
Clone this repository
-
Download image files according to the instructions in the Dataset section.
-
The notebooks can be run in any order, with the following exceptions:
4.1 The
baseline_experiments_results.ipynbnotebook will only reflect new runs if you run it afterward. You can view previously executed runs in that notebook, however.4.2 The
run_mmbt_masked_text_eval.ipynbandbertviz_attention.ipynbnotebooks runs on previously saved model(s); so you need to have trained and saved a model (MMBT and/or the BERT text-only) in the output directory specified in the--output_dirargument to successfully run this notebook.
4.2.1 the--output_dirarguments in these notebooks need to be the output directory where the model is saved; The easiest way to follow this is to make sure that the--output_dirargument for these notebooks are the same as the--output_dirargument in either therun_mmbt.ipynborrun_bert_text_only.ipynbnotebooks. -
To change hyperparameters for the text-only and MMBT notebooks, simply change the default values in the cell containing the Argument parser.
5.1 These notebooks can simply be run as is according to the default arguments.
5.2 To specify the experiment to be run, simply change the filenames of the desired datafile and specify the output directory, otherwise results will simply be written over the existing output directory. -
The loaded Tensorboard in the baseline_experiments_results.ipynb can be re-launched to reflect new experiment results.
We have created a shared Google Drive - LAP for this project where all the scripts, data, and Jupyter/Colab Notebooks in this repo have been uploaded. Simply download this drive and re-upload to your own Google Drive for testing. (i.e. you can only view files in this Drive and comment, but you cannot edit them.)
IMPORTANT: If you choose to test our project this way, please be aware that you still need to update the
path to the 'LAP' directory to reflect its location in your 'MyDrive'. Your pwd should always be the 'LAP'
project parent directory for the codes to work as intended. (The name for this directory needs not be 'LAP', but has
to be at the same level as described in the Directory Organization.)
Caveats: We reserve the ability to modify this Drive from time to time; particularly to manage storage. We may delete or write over files in this shared Drive without notice and access is only provided for your convenience. There may be log files from our experiments and previously run model weights; however those are not necessary for project reproducibility.
The notebooks in this directory contain the code to run the experiments. Please see each individual notebook for more detailed explanations. Using Google Colab is recommended since they were created and tested on Colab and running the models without GPU can take a long time. If you have access to a GPU outside of Colab, it is possible of course to run the experiments on an environment of your choice but the notebooks cannot be guaranteed to work on every possible setting.
-
baseline_experiments_results.ipynb shows the Tensorboard from the experiments with the textonly BERT model and the MMBT model
-
run_bert_text_only.ipynb shows the end-to-end pipeline for running the text-only experiments
-
run_mmbt.ipynb notebook shows the end-to-end MMBT experiment pipeline
-
bertviz_attention.ipynb shows the steps to visualize the attention mechanism using BertViz (Vig, 2019)
-
run_mmbt_masked_text_eval.ipynb shows the steps to test the MMBT model with masked text input for unimodal image classification
-
image_submodel.ipynb This notebook details the Image-only model and how we obtained our results from that experiment.
You can basically ignore the preprocess/ folder, since it's not relevant for running the any parts of the model. It includes various scripts that were used in extracting the labels from the dataset, checking that the created files match with the content of the reports and images they were created from and also filtering the frontal images from the rest.
These are experiments 1-3 listed in the Overview Section.
Although our Text-only model seems to perform slightly better than our multimodal model we were still curious to see if there exist some edge cases where the model actually benefits from multimodality. Indeed, we did find some of these edge cases where the text model does make a false predictions but MMBT predicts it correctly. The opposite is naturally true as well. There does exist other cases where MMBT makes a mistake and the text-model does not. This naturally follows from the fact that the text model achieves a higher accuracy. However, for this specific experiment we were more interested in the former case, where the model does benefit from multimodality in terms of making correct predictions. The results of this can be seen in the table below.
| Labeling Scheme | Total Errors (Text Model) | Total Corrected by MMBT | Corrected False Positives | Corrected False Negatives |
|---|---|---|---|---|
| Major | 17 | 3 | 0 | 3 |
| Impression | 70 | 31 | 12 | 19 |
We were curious to further investigate these cases where the text model makes a mistake and the multimodal model corrects it. Therefore, we decided to visualize the attention weights of some of these cases using the BertViz visualization tool by Jesse Vig (2019). We compared the attention weights of the text-only model to the attention weights of our multimodal model (MMBT) to see whether there are any visible differences between these cases. In the multimodal model, the image embeddings are projected into BERT's text token space and they are propagated through the self-attention layers, just like the normal text embeddings. This allows us to look at the attention weights of these image embeddings and see whether they attend more or less to other tokens and whether these connections can reveal anything about what leads the multimodal model to correct some of these cases where the text model fails.
In addition, visualizing attention weights can help reveal whether there are any general patterns that explain how the attention mechanism contributes to making predictions.
This constitutes experiment 4 in the Overview Section.
One additional experiment we conducted was to use our multimodal model on unseen unimodal images. Since our image model is the weak link in our multimodal setting, with clearly the lowest accuracy, we were curious to see whether using our finetuned multimodal model in a scenario where we mask all the text input and rely solely on the image modality would lead to a better performance than our image only model achieves.
This is experiment 5 in the Overview Section.
The integrated gradients is a way to visualize and extract explanations from the images. The basic idea behind it is that we can make use of the learned weights, which allow us to take the partial derivatives w/ respect to the inputs (the pixels) and visualize gradients that have highest activations with respect to some threshold value. The integrated gradients module is a fork from this repository https://github.com/TianhongDai/integrated-gradient-pytorch and it comes with an open source MIT license. We have slightly modified the original implementation to work with our data. For more information consult the original paper "Axiomatic Attribution for Deep Networks". Also consult the image_submodel.ipynb notebook for more details on how it was used in our experiment.
Otherwise, the integrated_gradients/ directory itself can be safely ignored in terms of running the experiments.
We evaluated our model on 2 different labeling schemes. In our main labeling scheme we extracted the labels from the 'major' field in the radiologists report. We labeled everything tagged as 'normal' as 0 and everything else as 'abnormal' or 1. This led to approximately 40% normal cases and 60% abnormal cases.
In addition, we experimented using the 'impression' field in the radiologists report as the basis for our labels,
as this is likely what a medical professional would look at in determining the significance of the report.
This led to approximately 60% normal cases and 40% abnormal cases, the exact opposite of our main labeling scheme.
We used the text from the 'findings' field in the radiologists report as our text modality for all of our experiments.
We then evaluated our models on both of these labeling schemes.
The results can be seen in the table below.
| Model | MMBT (major) | MMBT (impression) | Masked Text MMBT (major) | Masked Text MMBT (impression) |
|---|---|---|---|---|
| Multimodal | 0.96 | 0.86 | 0.66 | 0.77 |
| Text-only | 0.97 | 0.88 | N/A | N/A |
| Image-only | 0.74 | 0.74 | N/A | N/A |
In addition, we tried a third multilabel labeling scheme, which was based on the 'major' labels. However, we created a third category for cases that were borderline abnormal and did not involve any kinds of diseases, but other abnormalities, such as medical instruments or devices. We report our multilabel results in F1 instead of accuracy. The results can be seen in the table below.
| Model | MMBT - Binary (accuracy) | MMBT - Multilabel (macro/micro F1) | Masked Text MMBT - Binary (accuracy) | MMBT - Multilabel (macro/micro F1) |
|---|---|---|---|---|
| Multimodal | 0.96 | 0.82/0.93 | 0.66 | 0.41/0.55 |
| Text-only | 0.97 | 0.86/0.94 | N/A | N/A |
| Image-only | 0.74 | N/A | N/A | N/A |
From the results table in Multimodal Corrections, it is apparent that there are cases where the text-only model makes false predictions when the MMBT model does not. We selected an example to visualize the attention heads for comparison between the text-only and the MMBT model.
Sample ID: CXR2363
The last layer’s attention head of the text-only model, which falsely predicts the label for this sample.
The lines show the attention scores corresponding to the text tokens with line intensity being proportional to
attention weight. Tokens with higher attention scores are circled.

The last layer’s attention head of the MMBT model, which correctly predicts the label for this sample.
left figure = image embedding token 1,center = image embedding token 2, right = imageembedding token 3.
Tokens with higher attention scores are circled.
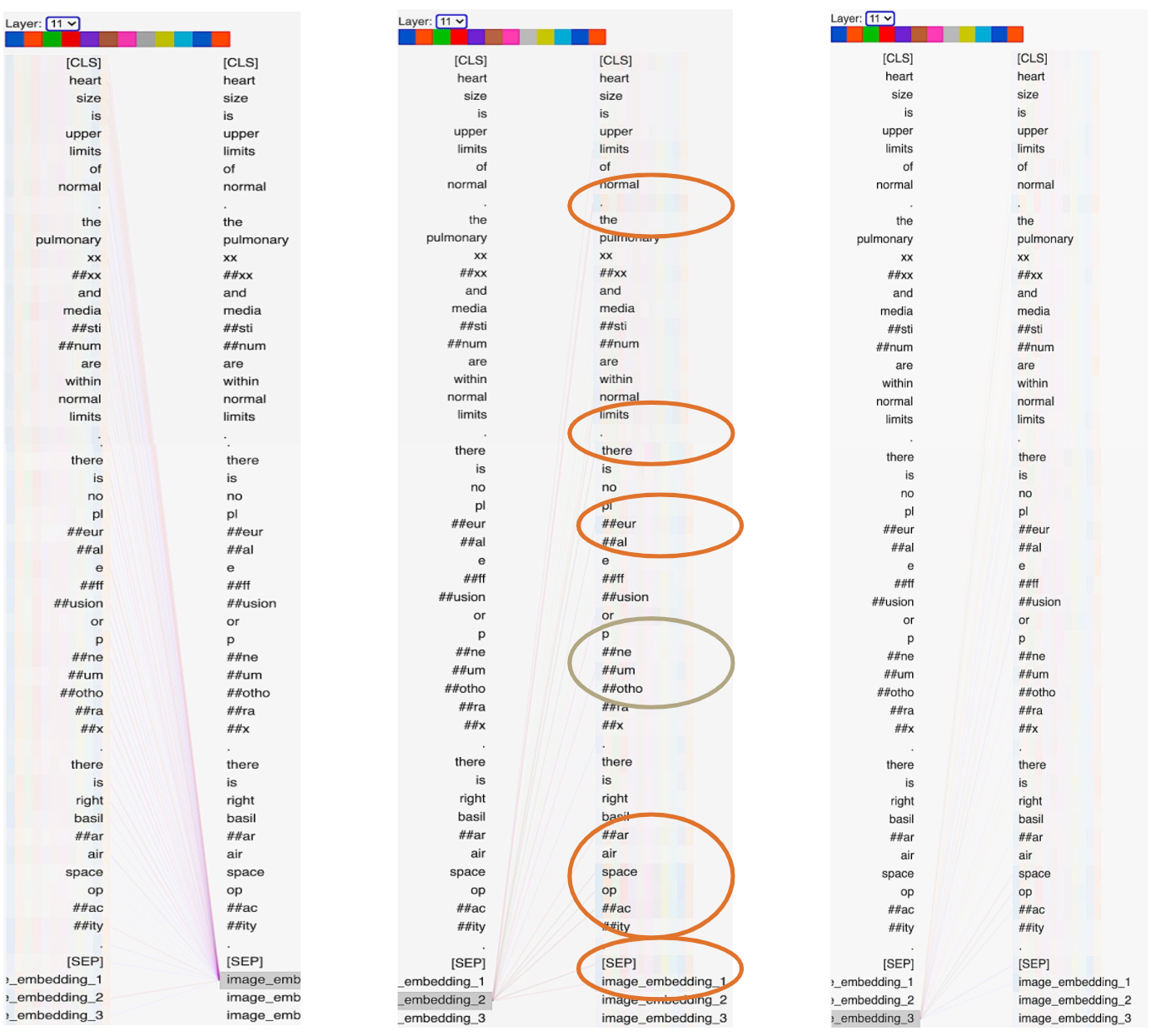
As can be examined in the images above, the MMBT model’s attention mechanism aligns the second image embedding token to content word tokens such as ‘pleural’, ‘pneumothorax’, ‘basilar’, ‘air’, ‘space’, and ‘opacity’ in addition to the ‘[SEP]’ and first image embedding token. In contrast, the attention mechanism in the text-only model aligns the ‘[CLS]’ token more strongly (higher attention scores) with the following tokens: ‘heart’, ‘is’, ‘there’, ‘no’, ‘.’, and ‘right’.
Results from this experiment are reported in the Masked Text columns of the tables in Findings: Unimodal vs Multimodal and Findings:Findings: Binary vs. Multilabel.
MMBT's improvement over the unimodal image-only model in this scenario is only observed when tested on the 'impression' labeled dataset.
Faik Aydin, Maggie Zhang, Michelle Ananda-Rajah, and Gholamreza Haffari. 2019. Medical Multimodal Classifiers Under Scarce Data Condition. arXiv:1902.08888.
Guillem Collell, Ted Zhang, and Marie-Francine Moens. 2017. Imagined visual representations as multimodal embeddings. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence, AAAI’17, pages 4378–4384, San Francisco, California, USA. AAAI Press.AAAI Press
Dina Demner-Fushman, Marc D. Kohli, Marc B. Rosenman, Sonya E. Shooshan, Laritza Rodriguez, Sameer Antani, George R. Thoma, and Clement J. McDonald. 2016. Preparing a collection of radiology examinations for distribution and retrieval. Journal of the American Medical Informatics Association : JAMIA, 23(2):304–310. DOI: 10.1093/jamia/ocv080
Jacob Devlin, Ming-Wei Chang, Kenton Lee, and Kristina Toutanova. 2019. BERT: Pre-training of Deep Bidirectional Transformers for Language Understanding. arXiv:1810.04805.
Allyson Ettinger. 2020. What BERT Is Not: Lessons from a New Suite of Psycholinguistic Diagnostics for Language Models. Transactions of the Association for Computational Linguistics, 8:34–48. arXiv:1907.13528
Kaiming He, Xiangyu Zhang, Shaoqing Ren, and Jian Sun. 2015. Deep Residual Learning for Image Recognition. arXiv:1512.03385.
Douwe Kiela, Suvrat Bhooshan, Hamed Firooz, Ethan Perez, and Davide Testuggine. 2020. Supervised Multimodal Bitransformers for Classifying Images and Text. arXiv:1909.02950.
Yoon Kim. 2014. Convolutional Neural Networks for Sentence Classification. arXiv:1408.5882.
Olga Kovaleva, Alexey Romanov, Anna Rogers, and Anna Rumshisky. 2019. Revealing the Dark Secrets of BERT. In Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP), pages 4365–4374, Hong Kong, China. Association for Computational Linguistics. arXiv:1908.08593
Jiasen Lu, Dhruv Batra, Devi Parikh, and Stefan Lee. 2019. ViLBERT: Pretraining Task-Agnostic Visiolinguistic Representations for Vision-and-Language Tasks. arXiv:1908.02265.
Pranav Rajpurkar, Jeremy Irvin, Kaylie Zhu, Brandon Yang, Hershel Mehta, Tony Duan, Daisy Ding, Aarti Bagul, Curtis Langlotz, Katie Shpanskaya, Matthew P. Lungren, and Andrew Y. Ng. 2017. CheXNet: Radiologist-Level Pneumonia Detection on Chest X-Rays with Deep Learning. arXiv:1711.05225.
Claudia Schulz and Damir Juric. 2020. Can Embeddings Adequately Represent Medical Terminology? New Large-Scale Medical Term Similarity Datasets Have the Answer! Proceedings of the AAAI Conference on Artificial Intelligence, 34(05):8775–8782. arXiv:2003.11082
Francesca Strik Lievers and Bodo Winter. 2018. Sensory language across lexical categories. Lingua, 204:45–61. https://doi.org/10.1016/j.lingua.2017.11.002
Mukund Sundararajan, Ankur Taly, Qiqi Yan. 2017. Axiomatic Attribution for Deep Networks. ArXiv:1703.01365.
Hao Tan and Mohit Bansal. 2019. LXMERT: Learning Cross-Modality Encoder Representations from Transformers. arXiv:1908.07490.
Jesse Vig. 2019. A Multiscale Visualization of Attention in the Transformer Model. In Proceedings of the 57th Annual Meeting of the Association for Computational Linguistics: System Demonstrations, pages 37–42, Florence, Italy. Association for Computational Linguistics. arXiv:1906.05714; BertViz GitHub
Xiaosong Wang, Yifan Peng, Le Lu, Zhiyong Lu, Mohammadhadi Bagheri, and Ronald M. Summers. 2017. ChestX-ray8: Hospital-scale Chest X-ray Database and Benchmarks on Weakly-Supervised Classification and Localization of Common Thorax Diseases. 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pages 3462–3471. arXiv:1705.02315