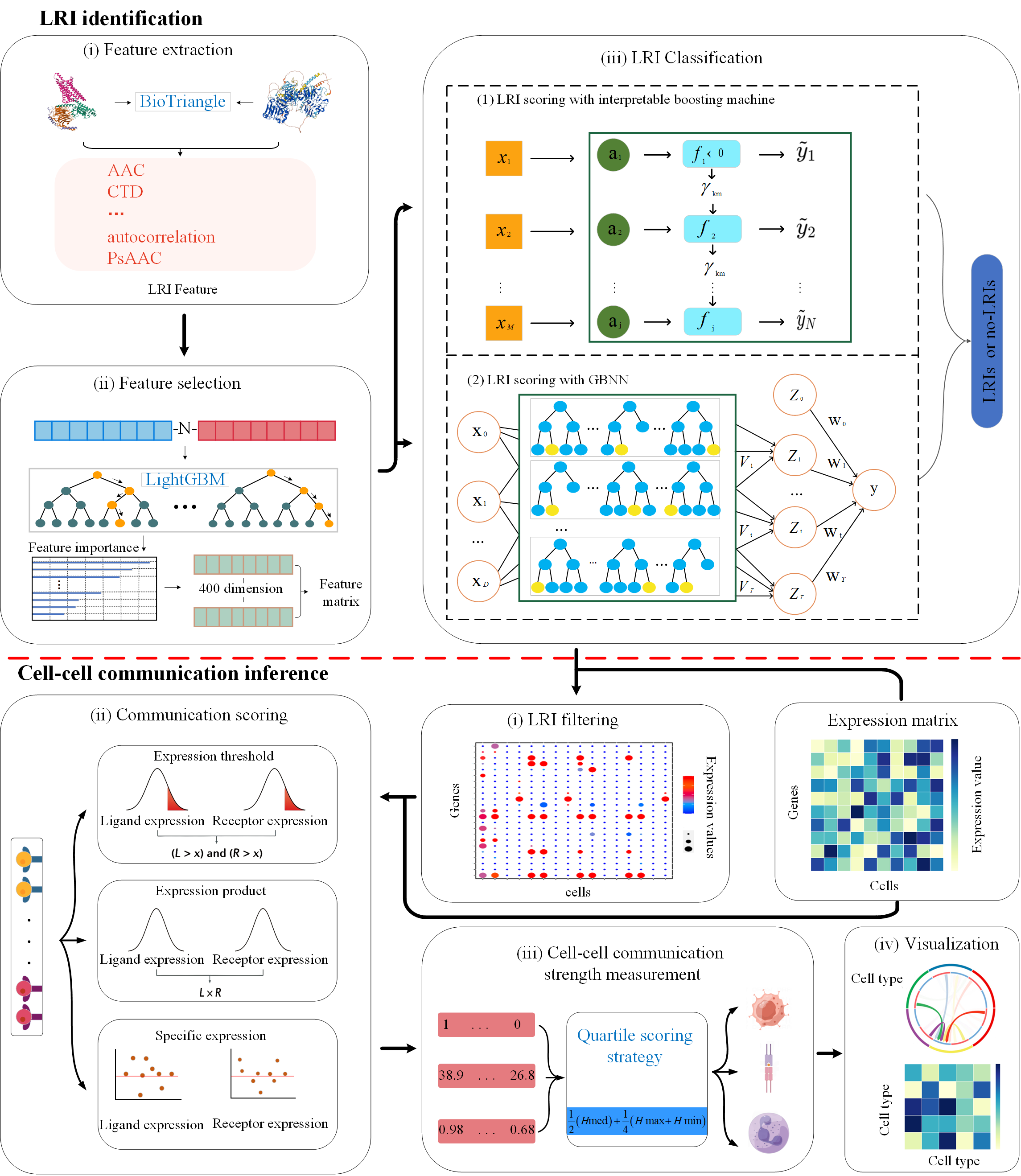
CellGiQ, a novel framework for deciphering ligand-receptor-mediated cell-cell communication by incorporating machine learning and a quartile scoring strategy from single-cell RNA sequencing data. CellGiQ accurately inferred intercellular communication within human HNSCC tissues. CellGiQ is anticipated to dissect cellular crosstalk and signal pathways at single cell resolution.
- python == 3.8.13
-
tensorflow == 2.10.0
-
keras == 2.10.0
-
GBNN == 0.0.2
-
interpret == 0.2.7
-
scikit-learn == 0.24.0
-
lightgbm == 3.3.5
-
wheel == 0.37.1
-
pands == 1.5.0
-
numpy == 1.24.2
1.Data is available at uniprot, GEO.
2.Feature extraction website at BioTriangle
-
We obtain ligand and receptor feature at BioTriangle
-
Run the model to obtain the LRI, or the user-specified LRI database
python code/CellGiQ.py -
Using quartile method (including Expression thresholding, Expression product and Specific expression), the cell-cell communication matrix was finally obtained.
python code/case study
If you want to test other tumors, just replace GSE103322.csv in the code case_study.py (Note: use the specified database to replace the datasetLRI_dataset.csv)