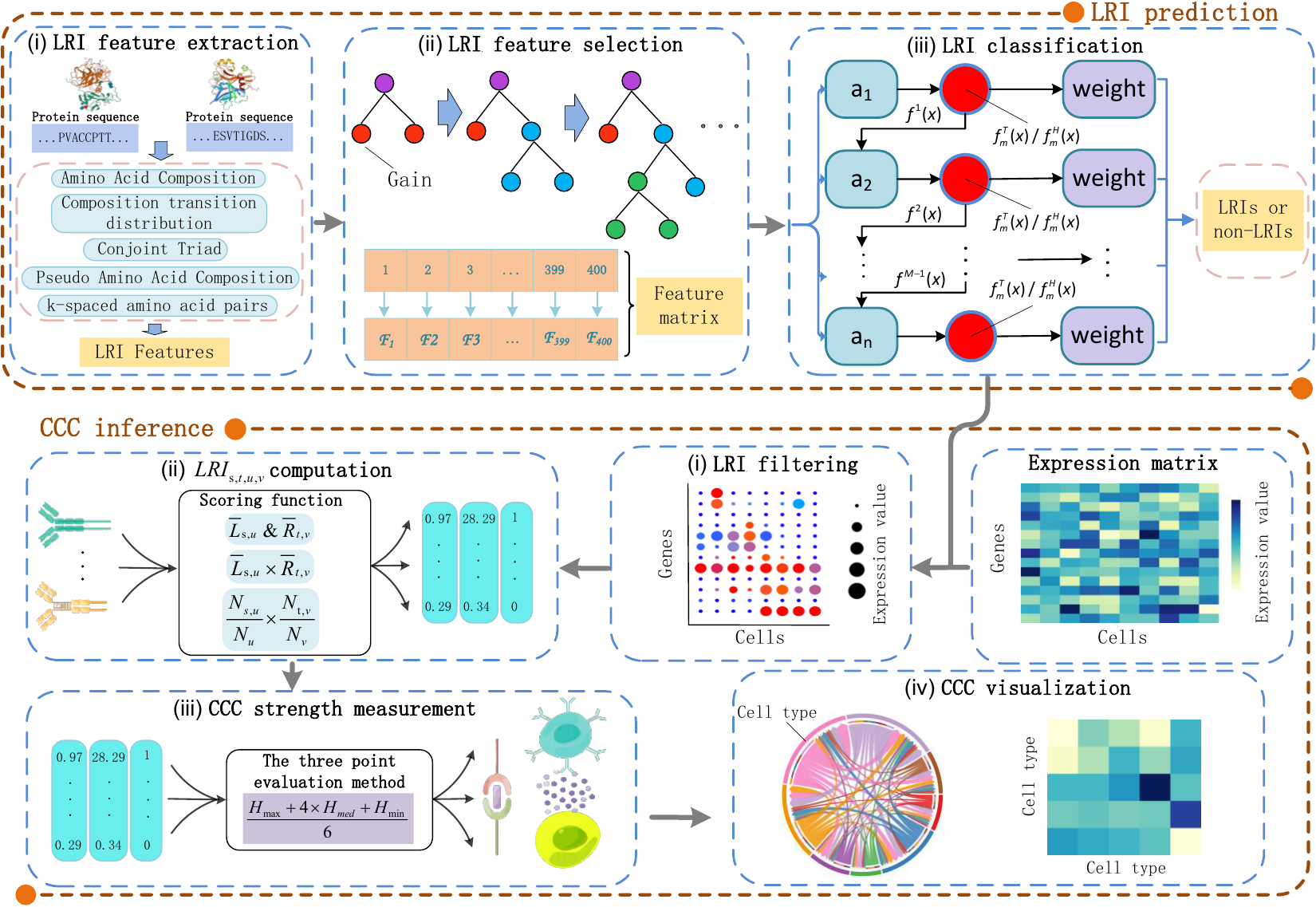
-
Single-cell RNA sequencing data can be downloaded from the GEO database. (melanoma (accession code:GSE72056), head and neck squamous cell carcinomas (HNSCC) (accession code: GSE103322), and colorectal cancer (CRC) (accession code: GSE81861)).
Install Python 3.9.15 for running this code. And these packages should be satisfied:
- seaborn >= 0.12.0
- pandas >= 1.5.3
- numpy >= 1.25.2
- scikit-learn >= 1.2.1
- matplotlib >= 3.7.2
- xgboost == 1.6.2
- lightgbm == 3.3.0
- KTBoost == 0.2.2
- gpboost == 0.7.10
CellPhoneDB, NATMI, iTALK, CellChat, CellComNet, CellEnBoost
See a Basic tutorial how to Use a Cell-Cell Communication Analysis Computing Framework Based on Ligand Receptor and Single Cell RNA Sequencing Data (CellDialog).
The tutorial with the test data takes minutes to complete!
The following is also a brief introduction to Usage.
- scRNA-seq data: Each column is a normalised gene/protein expression profile of a cell type or an individual cell. An example snapshot of the abundance matrix is shown below. And the scRNA-seq data was processed by code/Processing_scRNA-seq_data.py
| cell_type_0 | cell_type_1 | cell_type_2 | ... | |
|---|---|---|---|---|
| Gene1 | 1.56 | 2.84 | 53 | ... |
| Gene2 | 13 | 3.2 | 42.4 | ... |
| Gene3 | 0.245 | 0 | 0.66 | ... |
| ... | ... | ... | ... | ... |
- First, run the model, the default 5 fold cross-validation, get LRI pairs. Or the user can user-specified LRI database directly, skip this step to the second step.
python code/CellDialog.py
- The second step, Run the three-point estimation method, including [(cell expression)(expression product)(expression thresholding)]. (Note: the user-specified database only needs to replace the LRI.csv file and the corresponding format in the file.)
- Ligand-Receptor Interactions :The interaction data should be represented as a two-column table, with the first column containing ligands and the second column containing receptors (as in the following example).
| Ligand | Receptor |
|---|---|
| LIGAND1 | RECEPTOR1 |
| LIGAND2 | RECEPTOR2 |
| LIGAND3 | RECEPTOR2 |
| LIGAND4 | RECEPTOR3 |
| LIGAND4 | RECEPTOR4 |
| ... | ... |
python example/The_three-point_estimation_method.py
- Finally, output the strength of cell-cell communication.
This code was run on a computer with the following specifications:
- CPU: AMD EPYC 7302
- Memory: 256GB DDR4
- GPU: GeForce RTX 2080 Ti
However, the minimal requirements for running this protocol are:
- CPU: Intel Core i7-11800H
- Memory: 16GB DDR4
- GPU: NVIDIA GeForce RTX 3050Ti
- Storage: At least 10GB available
| CellDialog | CellPhoneDB | NATMI | iTALK | CellChat | CellComNet | CellEnBoost | |
|---|---|---|---|---|---|---|---|
| Time (S) | 194.72 | 85.56 | 35.22 | 10.31 | 183.04 | 1366.07 | 4104.3 |
| Memory (MB) | 164.89 | 387.64 | 102.7 | 724.42 | 1186.59 | 166.65 | 186.82 |