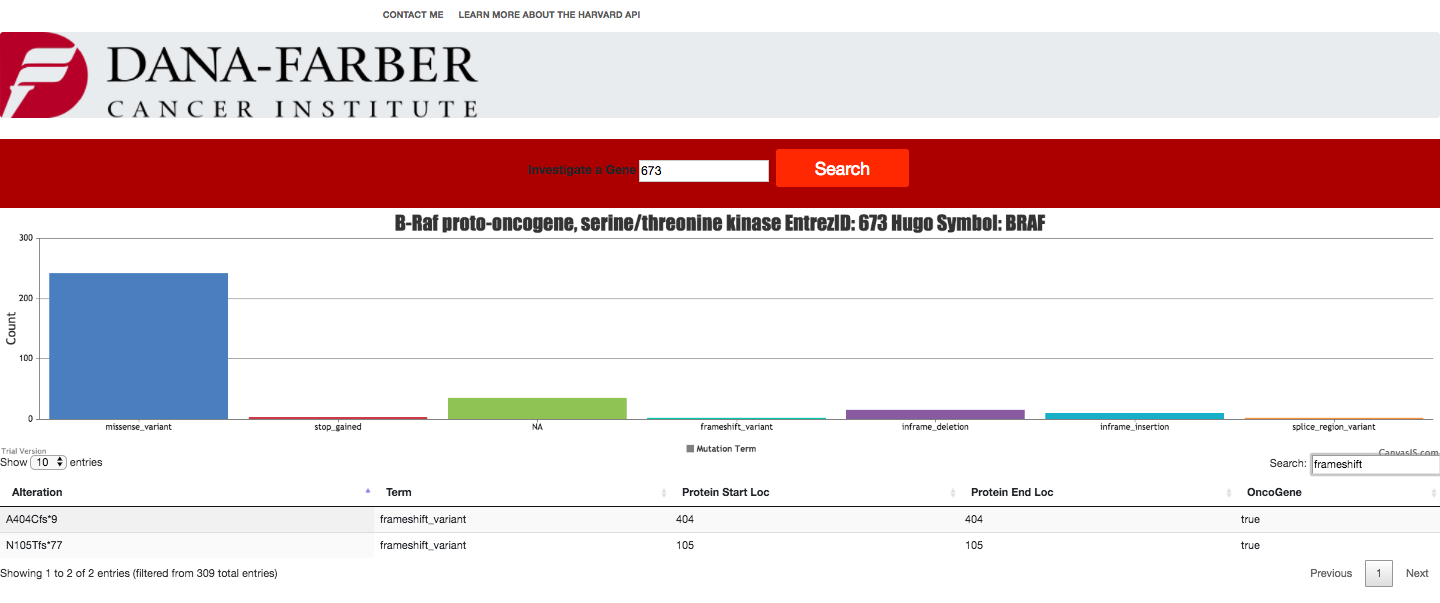
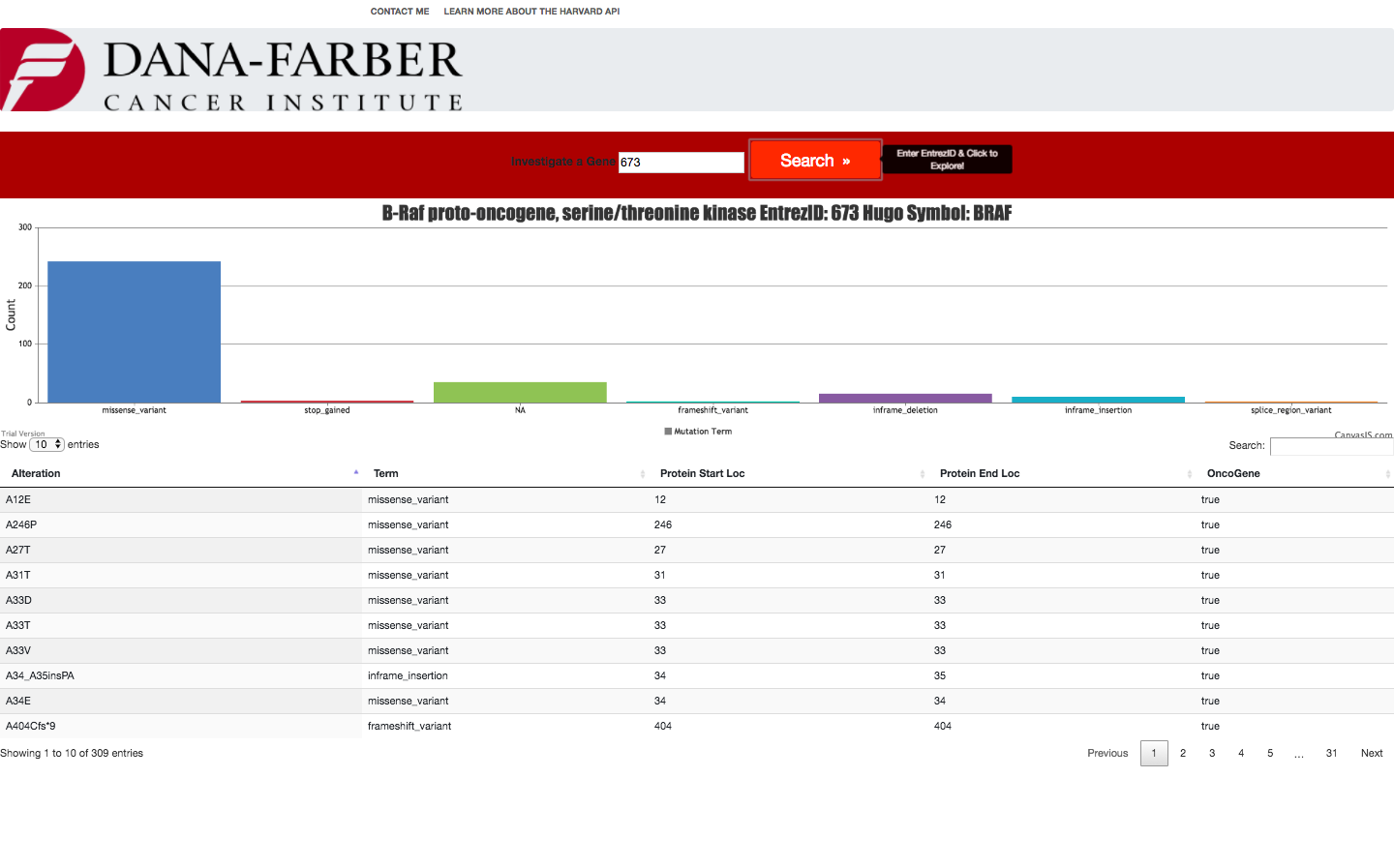
- Gene table must contain the following columns a. Alteration b. Hugo Symbol c. Entrez Gene Id d. OncoGene and or TSG i. Only when one or both terms are true
I did not include the desired part 1c or b in the table but the chart as each alteration of the given gene have a shared name, hugo symbol, and entrez gene id and thus the columns were redundant. Indeed, request 1d is redundant as well.
- Gene breakdown feature should have a bar chart that displays a count of data elements broken down by consequence[term].
- The columns displayed are currently hard coded, but can be loaded on the fly and set as hidden or visible as desired
- Paging of large result sets are paged and can be changed by user to view more options
- You can use the included search box to search the text of the data table and filter
- sort through each table column