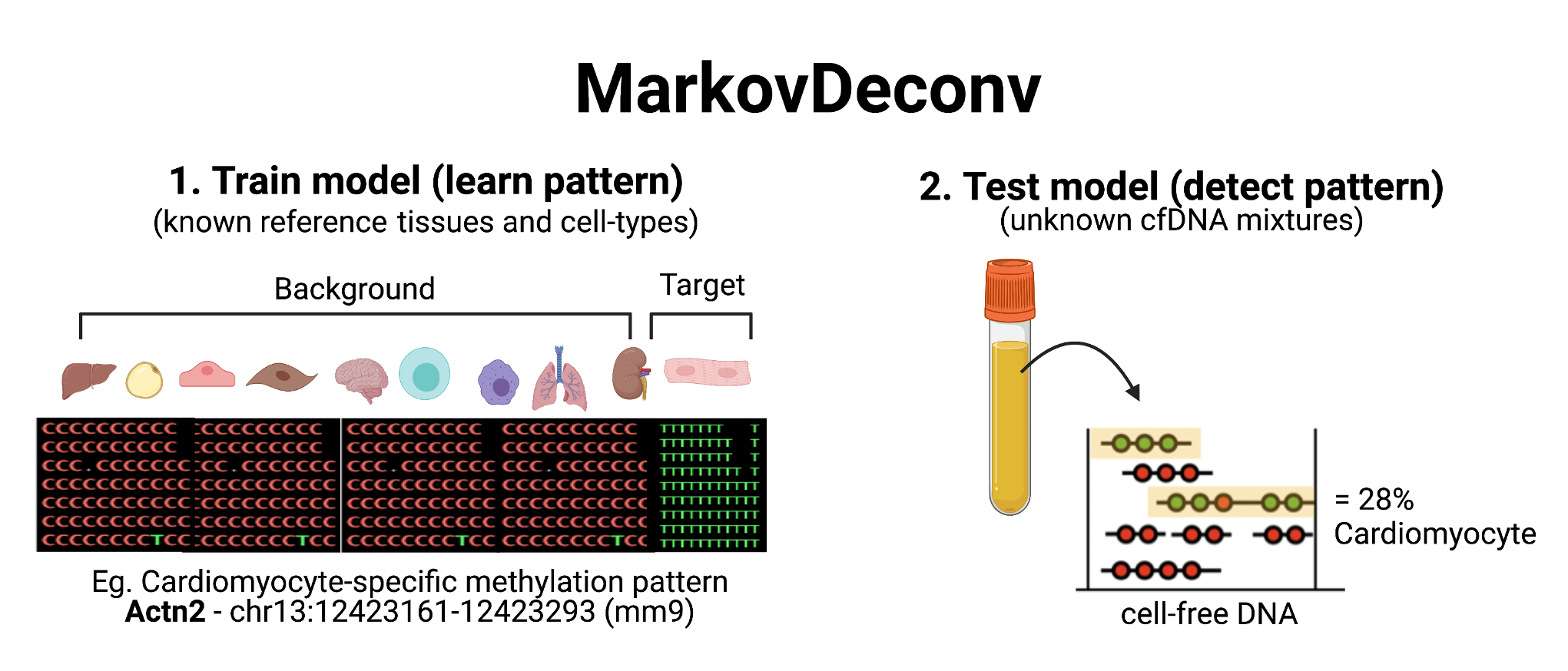
MarkovDeconv is a deconvolution model for DNA methylation sequencing data, designed to classify the cellular origins of cell-free DNA (cfDNA) fragments in blood samples. This ultrasensitive method uses fragment-level CpG methylation patterns to detect trace amounts of cell-type specific signals within complex cfDNA mixtures. Decoding the cellular origins of cfDNA from liquid biopsies serves as a promising new approach for non-invasive monitoring of tissue damage.
At identified cell-type specific DNA methylation patterns, this model is trained to recognize patterns belonging to cell-types of interest in unknown cfDNA mixtures. This allows for binary classification of each cfDNA molecule as belonging to the cell-type of interest or alternatively is classified as background.
First make sure you have wgbstools installed.
# Clone
git clone https://github.com/nloyfer/MarkovDeconv.git
cd MarkovDeconv/counter/
# compile
make
cd ..- python 3+ (tested with V3.8.10)
- samtools (tested with V1.12 using htslib V1.12)
- bedtools (tested with V2.27.1)
- boost c++ library (tested with V1.71.0)
MarkovDeconv requires only a standard computer with enough RAM to support the in-memory operations. For optimal performance, we recommend a computer with the following specs:
- RAM: 16+ GB
- CPU: 16 cores
This package is supported for macOS and Linux. The package has been tested on the following system:
- Linux: Ubuntu 20.04
- macOS: Catalina (10.15.7)
MarkovDeconv mainly depends on the Python scientific stack.
numpy
scipy
pandas
matplotlib
Now you can detect cfDNA fragments originating from cell-types of interest.
First, train the model to distinguish CpG Methylation patterns of the target cell-type from background.
This command takes as input:
- marker file: a
bedfile with 2 extra columns for CpG indexes. Could be the output of thewgbstools segmentcommand, or any custom bed file once you added the [startCpG, endCpG] columns withwgbstools convert -L BED_FILE. - group file: a
csvtable\ text file defining which pat files are target (group1) and which are background (group2) - pat files: a set of pat files from known reference cell-types to train the model. You can generate
patfiles out ofbamfiles for each of the reference cell-types using thewgbstoolsbam2patcommand.
python train.py markers.bed -g groups.csv -f -v -o ./my_train_dir --reference_data /path/to/reference/gDNA/files/Then, deconvolve unknown cfDNA mixtures to identify molecules originating from the target cell-type
python deconvolve.py /path/to/my_train_dir/ -v --target TARGET-CELLTYPE --pats /path/to/test/cfDNA/files/*pat.gzThis tutorial comes with step-by-step details about how to use the MarkovDeconv model that should take ~10 minutes on a recommended computer.