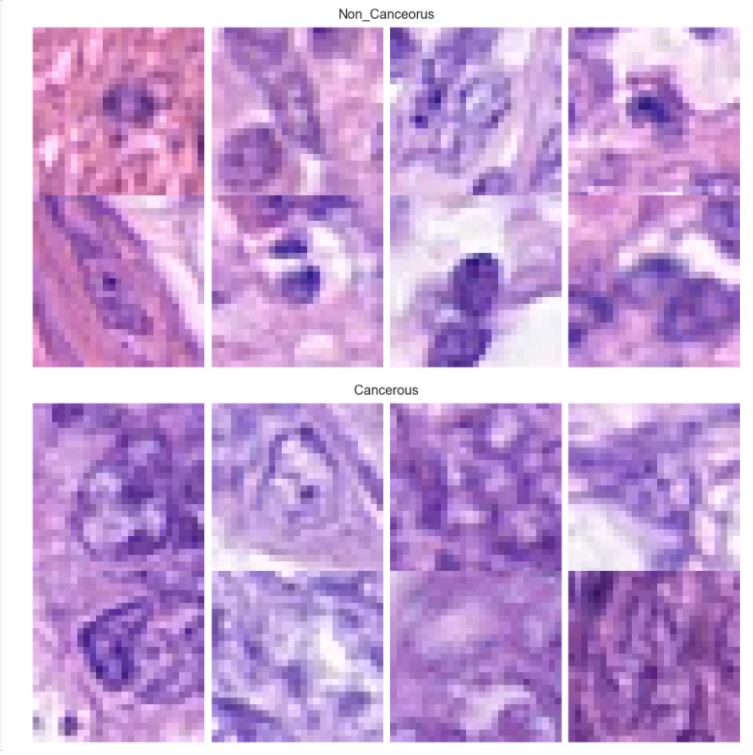
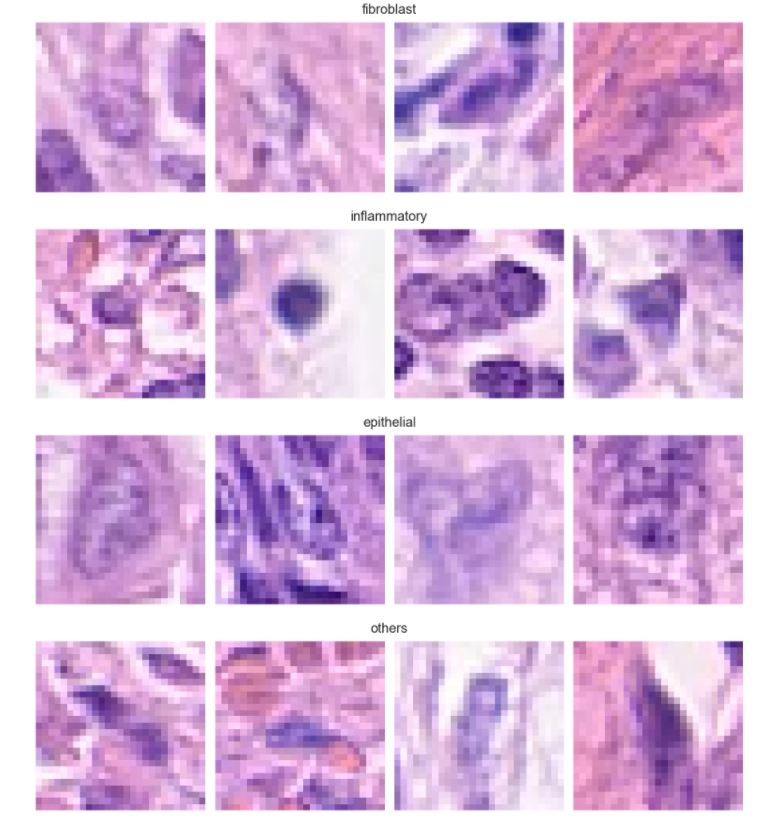
Detection of Colon Cancer and its Cell Types using Histopathological Images: A deep learning approach
Colorectal cancer (colon cancer) is a type of chronic illness that affects the colon and rectum of an individual and is the cause of the second most cancer-related deaths in the US. Pathologists perform tissue-based diagnosis using a microscope to classify cell nuclei from Routine Colon Cancer images. Several CNN models like RCCNet, VGG-16 have demonstrated superior results in classifying colon cancer cells. In this project, we summarize our findings by analyzing several deep learning architectures to classify images according to cell type and whether the cell image represents a cancerous cell.
We used a modified version of the “CRCHistoPhenotypes” dataset for this project. The dataset consists of 27x27 RGB images of colon cells from 99 different patients. While inspecting the data, information from patientID 76 was missing resulting in histopathological images of 98 patients.
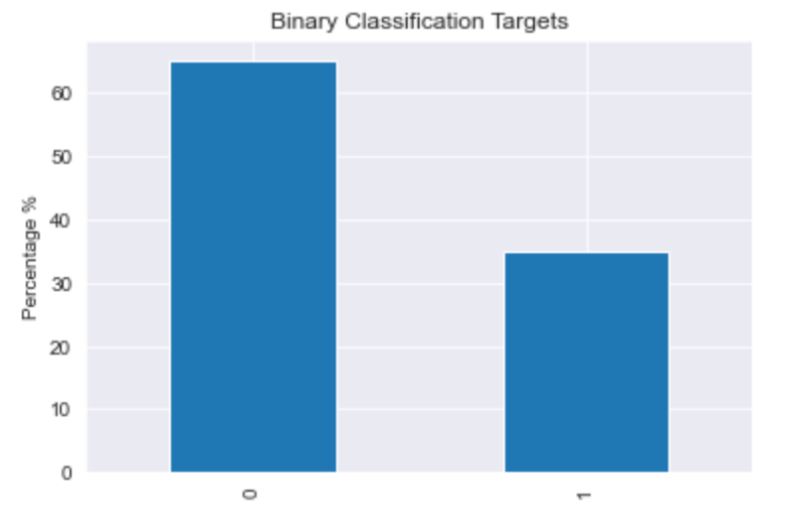
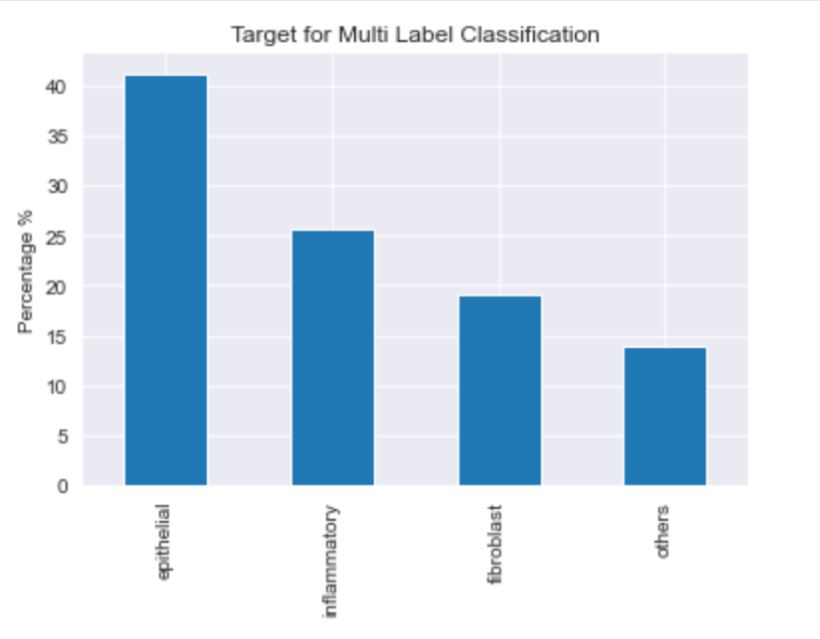
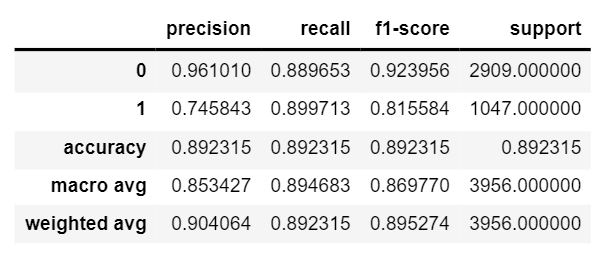
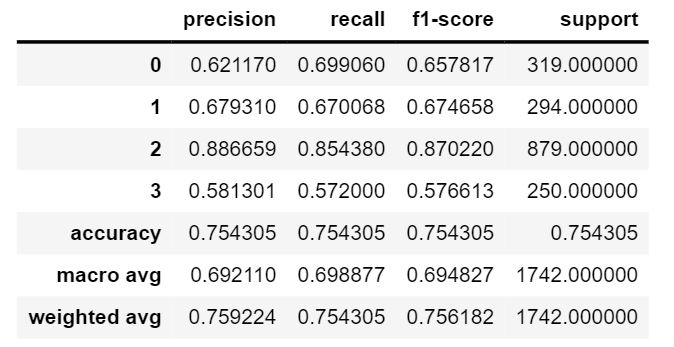
The dataset consists of 20280 images that have been classified as cancerous or non cancerous. Out of those, only 10284 images have a label for cell-type named as fibroblast, inflammatory, epithelial, and others, whereas 9996 images do not have a label for cell type. In figure 1, it can be seen that about 42% of the images belong to epithelial, 26% as inflammatory, 18% as fibroblast and 14% as others. In total, 33% of the images have been classified as “Cancerous” and the remaining 67% as “non-Cancerous”. This shows the dataset has unequal distribution of class labels and requires specialized metrics for model evaluation. We selected the macro average f1 score to give equal importance to all the class labels and assess the performance of both learning tasks. Additionally, classification metrics such as precision, recall and confusion matrices were analyzed to see how the model performed on each target label.
Data splitting was done in such a way that all samples from the same patientID will be in either train or test but not on both. This helps in training and testing the performance of the model more accurately and avoid any data leakage. We used a 4:1 ratio to create a training and independent test set. The training set was further split on a 4:1 ratio into training and validation sets. For processing images, we used Keras ImageDataGenerator and normalized the values between 0-1. In addition, we used data augmenting techniques such as rotations, shifts, horizontal and vertical flips to increase the nature of training samples and avoid any potential overfitting.
We developed our baseline model by using just two blocks of VGG architecture. We chose two blocks because of the small image size in the dataset whereas VGG architecture has been proposed to classify large size images. The blocks contains 64, 128 filters of size 3X3 followed by a pooling layer of 2X2 sizes respectively. The max pooling layer of 2X2 sizes are used to reduce feature dimensions by half. The feature map generated by the pool layer of the second block is flattened into a single feature vector. The last block takes the flattened input and produces two values as output using softmax activation. The baseline model trained on 100 epochs showed overfitting. We then tried data augmentation on the training set and added a ridge regularizer of 0.01 to apply a penalty on the layers kernel to fix overfitting.
| Method | #Parameters | #Test F1_Score |
|---|---|---|
| 2-Layer NN | 593,282 | 81.73 % |
| VGG 3 Blocks | 442,978 | 86.97 % |
We developed our baseline model by using just two blocks of VGG architecture. We chose two blocks because of the small image size in the dataset whereas VGG architecture has been proposed to classify large size images. The blocks contains 64, 128 filters of size 3X3 followed by a pooling layer of 2X2 sizes respectively. The max pooling layer of 2X2 sizes are used to reduce feature dimensions by half. The feature map generated by the pool layer of the second block is flattened into a single feature vector. Two dense layers of size 256 take the flattened input and produce four values as output using softmax activation. The baseline model trained on 100 epochs showed overfitting. We then tried decreasing the filter size on the dense layer to fix overfitting. The model tested on the test dataset showed great results compared to other VGG models.
| Method | #Parameters | #Test F1_Score |
|---|---|---|
| 2-Layer NN | 280,580 | 44.35 % |
| RCC Net | 690,212 | 57.26 % |
| Softmax CNN | 899,200 | 71.2 % |
| VGG 2 Blocks | 555,396 | 69 % |
We conducted several experiments to classify the images according to cell type and check if the image represents a cancerous cell. Initially, the CNN models were trained by adding a single hidden layer. The number of neurons was chosen in the power of 2, but greater than the number of classes. Gradually, we increased the number of hidden layers and number of neurons in the added layer. The performance of these models were analyzed and different techniques like regularization, drop out, early stopping and augmentation techniques were applied as necessary to reduce overfitting. For both cases, VGG architecture outperformed the rest of the models in our case. This experiment was limited to using base cnn architecture with a primary objective to analyse performance over a fully connected neural network. In future, we intend to extend this work using other CNN architectures and leverage pre-trained models for more accurate results.
The notebook containing exploratory data analysis is available here The notebook containing cancer cell detection is available here The notebook contaning cell type classification is available here