“From R to your manuscript”
easystats::install_easystats_latest()). Thanks for your support!
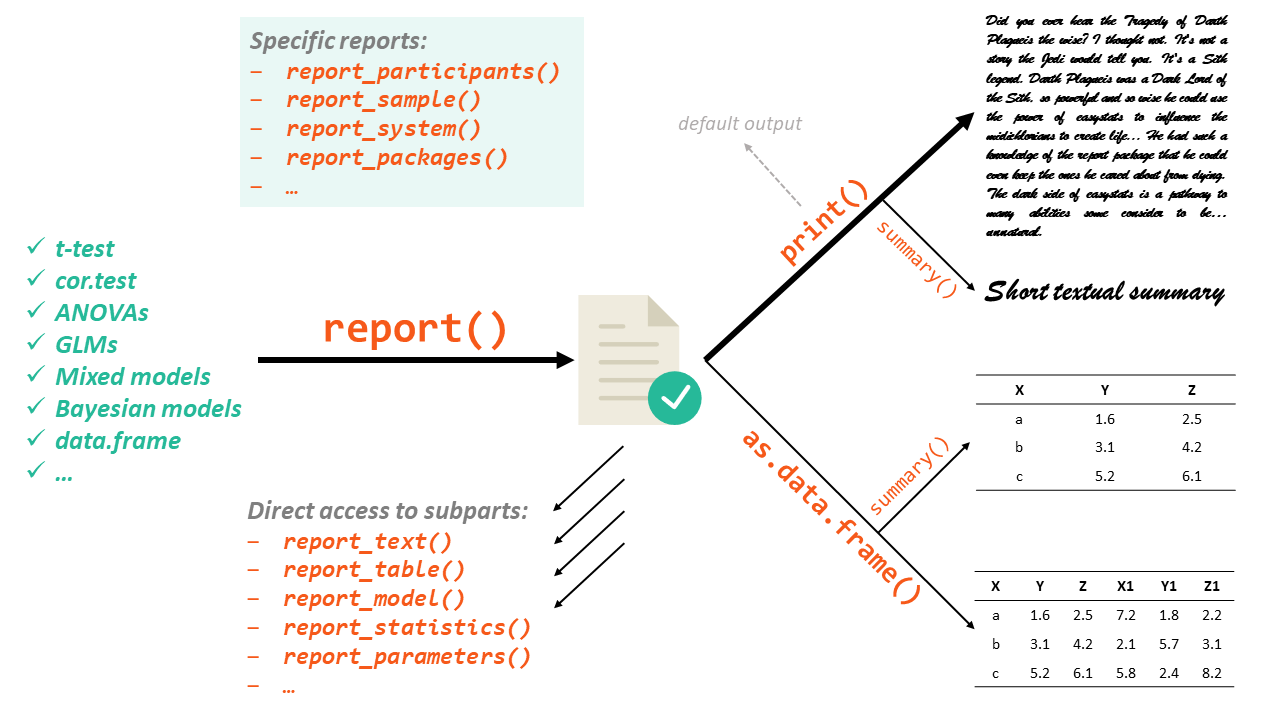
report’s primary goal is to bridge the gap between R’s output and the formatted results contained in your manuscript. It automatically produces reports of models and dataframes according to best practices guidelines (e.g., APA’s style), ensuring standardization and quality in results reporting.
library(report)
model <- lm(Sepal.Length ~ Species, data = iris)
report(model)# We fitted a linear model (estimated using OLS) to predict Sepal.Length with Species (formula: Sepal.Length ~
# Species). The model explains a significant and substantial proportion of variance (R2 = 0.62, F(2, 147) =
# 119.26, p < .001, adj. R2 = 0.61). The model's intercept, corresponding to Species = setosa, is at 5.01 (95%
# CI [4.86, 5.15], t(147) = 68.76, p < .001). Within this model:
#
# - The effect of Species [versicolor] is significantly positive (beta = 0.93, 95% CI [0.73, 1.13], t(147) =
# 9.03, p < .001; Std. beta = 1.12, 95% CI [0.88, 1.37])
# - The effect of Species [virginica] is significantly positive (beta = 1.58, 95% CI [1.38, 1.79], t(147) =
# 15.37, p < .001; Std. beta = 1.91, 95% CI [1.66, 2.16])
#
# Standardized parameters were obtained by fitting the model on a standardized version of the dataset.
The package documentation can be found here. Check-out these tutorials:
report is a young package in need of affection. You can easily be a part of the developing community of this open-source software and improve science! Don’t be shy, try to code and submit a pull request (See the contributing guide). Even if it’s not perfect, we will help you make it great!
Run the following:
install.packages("remotes")
remotes::install_github("easystats/report") # You only need to do that oncelibrary("report") # Load the package every time you start RThe report package works in a two step fashion. First, you create a
report object with the report() function. Then, this report object
can be displayed either textually (the default output) or as a table,
using as.data.frame(). Moreover, you can also access a more digest and
compact version of the report using summary() on the report object.
The report() function works on a variety of models, as well as other
objects such as dataframes:
report(iris)# The data contains 150 observations of the following variables:
# - Sepal.Length: n = 150, Mean = 5.84, SD = 0.83, Median = 5.80, MAD = 1.04, range: [4.30, 7.90], Skewness =
# 0.31, Kurtosis = -0.55, 0% missing
# - Sepal.Width: n = 150, Mean = 3.06, SD = 0.44, Median = 3.00, MAD = 0.44, range: [2, 4.40], Skewness = 0.32,
# Kurtosis = 0.23, 0% missing
# - Petal.Length: n = 150, Mean = 3.76, SD = 1.77, Median = 4.35, MAD = 1.85, range: [1, 6.90], Skewness =
# -0.27, Kurtosis = -1.40, 0% missing
# - Petal.Width: n = 150, Mean = 1.20, SD = 0.76, Median = 1.30, MAD = 1.04, range: [0.10, 2.50], Skewness =
# -0.10, Kurtosis = -1.34, 0% missing
# - Species: 3 levels, namely setosa (n = 50, 33.33%), versicolor (n = 50, 33.33%) and virginica (n = 50,
# 33.33%)
These reports nicely work within the tidyverse workflow:
library(dplyr)
iris %>%
select(-starts_with("Sepal")) %>%
group_by(Species) %>%
report() %>%
summary()# The data contains 150 observations, grouped by Species, of the following variables:
#
# - setosa (n = 50):
# - Petal.Length: Mean = 1.46, SD = 0.17, range: [1, 1.90]
# - Petal.Width: Mean = 0.25, SD = 0.11, range: [0.10, 0.60]
#
# - versicolor (n = 50):
# - Petal.Length: Mean = 4.26, SD = 0.47, range: [3, 5.10]
# - Petal.Width: Mean = 1.33, SD = 0.20, range: [1, 1.80]
#
# - virginica (n = 50):
# - Petal.Length: Mean = 5.55, SD = 0.55, range: [4.50, 6.90]
# - Petal.Width: Mean = 2.03, SD = 0.27, range: [1.40, 2.50]
Reports can be used to automatically format tests like t-tests or correlations.
report(t.test(mtcars$mpg ~ mtcars$am))# Effect sizes were labelled following Cohen's (1988) recommendations.
#
# The Welch Two Sample t-test testing the difference of mtcars$mpg by mtcars$am (mean in group 0 = 17.15, mean
# in group 1 = 24.39) suggests that the effect is positive, significant and large (difference = 7.24, 95% CI
# [-11.28, -3.21], t(18.33) = -3.77, p < .01; Cohen's d = -1.41, 95% CI [-2.17, -0.51])
As mentioned, you can also create tables with the as.data.frame()
functions, like for example with this correlation test:
cor.test(iris$Sepal.Length, iris$Sepal.Width) %>%
report() %>%
as.data.frame()
# Parameter1 | Parameter2 | r | 95% CI | t(148) | p | Method
# --------------------------------------------------------------------------------------------------------------------
# iris$Sepal.Length | iris$Sepal.Width | -0.12 | [-0.27, 0.04] | -1.44 | 0.152 | Pearson's product-moment correlationThis works great with ANOVAs, as it includes effect sizes and their interpretation.
aov(Sepal.Length ~ Species, data = iris) %>%
report()# The ANOVA (formula: Sepal.Length ~ Species) suggests that:
#
# - The main effect of Species is significant and large (F(2, 147) = 119.26, p < .001; Eta2 = 0.62, 90% CI
# [0.54, 0.68])
#
# Effect sizes were labelled following Field's (2013) recommendations.
Reports are also compatible with GLMs, such as this logistic regression:
model <- glm(vs ~ mpg * drat, data = mtcars, family = "binomial")
report(model)# We fitted a logistic model (estimated using ML) to predict vs with mpg and drat (formula: vs ~ mpg * drat).
# The model's explanatory power is substantial (Tjur's R2 = 0.51). The model's intercept, corresponding to mpg
# = 0 and drat = 0, is at -33.43 (95% CI [-77.90, 3.25], p = 0.083). Within this model:
#
# - The effect of mpg is non-significantly positive (beta = 1.79, 95% CI [-0.10, 4.05], p = 0.066; Std. beta =
# 3.63, 95% CI [1.36, 7.50])
# - The effect of drat is non-significantly positive (beta = 5.96, 95% CI [-3.75, 16.26], p = 0.205; Std. beta
# = -0.36, 95% CI [-1.96, 0.98])
# - The interaction effect of drat on mpg is non-significantly negative (beta = -0.33, 95% CI [-0.83, 0.15], p
# = 0.141; Std. beta = -1.07, 95% CI [-2.66, 0.48])
#
# Standardized parameters were obtained by fitting the model on a standardized version of the dataset. 95%
# Confidence Intervals (CIs) and p-values were computed using
Mixed models (coming from example from the lme4 package), which
popularity and usage is exploding, can also be reported as it should:
library(lme4)
model <- lme4::lmer(Sepal.Length ~ Petal.Length + (1 | Species), data = iris)
report(model)# We fitted a linear mixed model (estimated using REML and nloptwrap optimizer) to predict Sepal.Length with
# Petal.Length (formula: Sepal.Length ~ Petal.Length). The model included Species as random effect (formula: ~1
# | Species). The model's total explanatory power is substantial (conditional R2 = 0.97) and the part related
# to the fixed effects alone (marginal R2) is of 0.66. The model's intercept, corresponding to Petal.Length =
# 0, is at 2.50 (95% CI [1.20, 3.81], t(146) = 3.75, p < .001). Within this model:
#
# - The effect of Petal.Length is significantly positive (beta = 0.89, 95% CI [0.76, 1.01], t(146) = 13.93, p <
# .001; Std. beta = 1.89, 95% CI [1.63, 2.16])
#
# Standardized parameters were obtained by fitting the model on a standardized version of the dataset. 95%
# Confidence Intervals (CIs) and p-values were computed using the Wald approximation.
Bayesian models can also be reported using the new SEXIT framework, that combines clarity, precision and usefulness.
library(rstanarm)
model <- stan_glm(mpg ~ qsec + wt, data = mtcars)
report(model)# We fitted a Bayesian linear model (estimated using MCMC sampling with 4 chains of 1000 iterations and a
# warmup of 500) to predict mpg with qsec and wt (formula: mpg ~ qsec + wt). Priors over parameters were set as
# normal (mean = 0.00, SD = 8.43) and normal (mean = 0.00, SD = 15.40) distributions. The model's explanatory
# power is substantial (R2 = 0.81, 89% CI [0.73, 0.88], adj. R2 = 0.78). The model's intercept, corresponding
# to qsec = 0 and wt = 0, is at 19.87 (95% CI [9.27, 30.28]). Within this model:
#
# - The effect of qsec (Median = 0.92, 0.95% CI [0.40, 1.48]) has a 99.80% probability of being positive (> 0),
# 99.00% of being significant (> 0.30), and 0.15% of being large (> 1.81). The estimation successfuly converged
# (Rhat = 1.000) and the indices are reliable (ESS = 1682)
# - The effect of wt (Median = -5.03, 0.95% CI [-6.00, -4.12]) has a 100.00% probability of being negative (<
# 0), 100.00% of being significant (< -0.30), and 100.00% of being large (< -1.81). The estimation successfuly
# converged (Rhat = 0.999) and the indices are reliable (ESS = 2111)
#
# Following the Sequential Effect eXistence and sIgnificance Testing (SEXIT) framework, we report the median of
# the posterior distribution and its 95% CI (Highest Density Interval), along the probability of direction
# (pd), the probability of significance and the probability of being large. The thresholds beyond which the
# effect is considered as significant (i.e., non-negligible) and large are |0.30| and |1.81| (corresponding
# respectively to 0.05 and 0.30 of the outcome's SD). Convergence and stability of the Bayesian sampling has
# been assessed using R-hat, which should be below 1.01 (Vehtari et al., 2019), and Effective Sample Size
# (ESS), which should be greater than 1000 (Burkner, 2017).
One can, for complex reports, directly access the pieces of the reports:
model <- lm(Sepal.Length ~ Species, data = iris)
report_model(model)
report_performance(model)
report_statistics(model)# linear model (estimated using OLS) to predict Sepal.Length with Species (formula: Sepal.Length ~ Species)
# The model explains a significant and substantial proportion of variance (R2 = 0.62, F(2, 147) = 119.26, p < .001, adj. R2 = 0.61)
# beta = 5.01, 95% CI [4.86, 5.15], t(147) = 68.76, p < .001; Std. beta = -1.01, 95% CI [-1.18, -0.84]
# beta = 0.93, 95% CI [0.73, 1.13], t(147) = 9.03, p < .001; Std. beta = 1.12, 95% CI [0.88, 1.37]
# beta = 1.58, 95% CI [1.38, 1.79], t(147) = 15.37, p < .001; Std. beta = 1.91, 95% CI [1.66, 2.16]
This can be useful to complete the Participants paragraph of your manuscript.
data <- data.frame(
"Age" = c(22, 23, 54, 21),
"Sex" = c("F", "F", "M", "M")
)
paste(
report_participants(data, spell_n = TRUE),
"were recruited in the study by means of torture and coercion."
)# [1] "Four participants (Mean age = 30.0, SD = 16.0, range: [21, 54]; 50.0% females) were recruited in the study by means of torture and coercion."
Report can also help you create sample description table (also referred to as Table 1).
| Variable | setosa (n=50) | versicolor (n=50) | virginica (n=50) | Total |
|---|---|---|---|---|
| Mean Sepal.Length (SD) | 5.01 (0.35) | 5.94 (0.52) | 6.59 (0.64) | 5.84 (0.83) |
| Mean Sepal.Width (SD) | 3.43 (0.38) | 2.77 (0.31) | 2.97 (0.32) | 3.06 (0.44) |
| Mean Petal.Length (SD) | 1.46 (0.17) | 4.26 (0.47) | 5.55 (0.55) | 3.76 (1.77) |
| Mean Petal.Width (SD) | 0.25 (0.11) | 1.33 (0.20) | 2.03 (0.27) | 1.20 (0.76) |
Finally, report includes some functions to help you write the data analysis paragraph about the tools used.
report(sessionInfo())# Analyses were conducted using the R Statistical language (version 4.0.3; R Core Team, 2020) on macOS Catalina
# 10.15.7, using the packages Rcpp (version 1.0.6; Dirk Eddelbuettel and Romain Francois, 2011), Matrix
# (version 1.2.18; Douglas Bates and Martin Maechler, 2019), lme4 (version 1.1.26; Douglas Bates et al., 2015),
# rstanarm (version 2.21.1; Goodrich B et al., 2020), dplyr (version 1.0.4; Hadley Wickham et al., 2021) and
# report (version 0.2.0; Makowski et al., 2020).
#
# References
# ----------
# - Dirk Eddelbuettel and Romain Francois (2011). Rcpp: Seamless R and C++ Integration. Journal of Statistical
# Software, 40(8), 1-18. URL https://www.jstatsoft.org/v40/i08/.
# - Douglas Bates and Martin Maechler (2019). Matrix: Sparse and Dense Matrix Classes and Methods. R package
# version 1.2-18. https://CRAN.R-project.org/package=Matrix
# - Douglas Bates, Martin Maechler, Ben Bolker, Steve Walker (2015). Fitting Linear Mixed-Effects Models Using
# lme4. Journal of Statistical Software, 67(1), 1-48. doi:10.18637/jss.v067.i01.
# - Goodrich B, Gabry J, Ali I & Brilleman S. (2020). rstanarm: Bayesian applied regression modeling via Stan.
# R package version 2.21.1 https://mc-stan.org/rstanarm.
# - Hadley Wickham, Romain François, Lionel Henry and Kirill Müller (2021). dplyr: A Grammar of Data
# Manipulation. R package version 1.0.4. https://CRAN.R-project.org/package=dplyr
# - Makowski, D., Lüdecke, D., & Ben-Shachar, M.S. (2020). Automated reporting as a practical tool to improve
# reproducibility and methodological best practices adoption. CRAN. Available from
# https://github.com/easystats/report. doi: .
# - R Core Team (2020). R: A language and environment for statistical computing. R Foundation for Statistical
# Computing, Vienna, Austria. URL https://www.R-project.org/.
If you like it, you can put a star on this repo, and cite the package as follows:
- Makowski, D., Ben-Shachar, M. S., & Lüdecke, D. (2020). Automated reporting as a practical tool to improve reproducibility and methodological best practices adoption. CRAN.