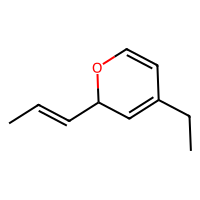
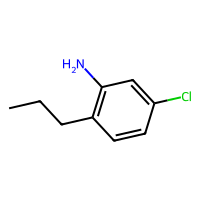
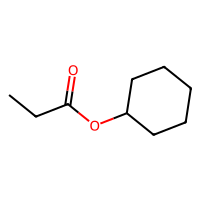
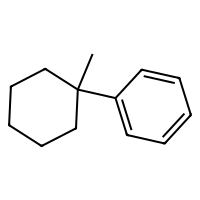
SELFIES are a robust and discrete representation of molecules1, which are a sort of successor to SMILES2. This repository contains a minimal working example of an MLP variational autoencoder that can be trained on SELFIES of at most 20 tokens, including how to download a database of SELFIES strings, how to process these as categorical data, the training loops, and some samples from the latent space (see above).
This code was written with Python 3.9 in mind. If you are using Conda, try
conda create -n minimal-vae-on-selfies python=3.9following with
pip install -r requirements.txtYou should add the src/ file to your PYTHONPATH. If you're using VSCode for running and debugging, this can be done by adding this key-value pair to your launch.json :
"env": {
"PYTHONPATH": "${workspaceFolder}${pathSeparator}src:${env:PYTHONPATH}"
}In src/data_preprocessing you can find files for downloading the dataset (which is PubChem's CID-SMILES saved at ftp:https://ftp.ncbi.nlm.nih.gov/pubchem/Compound/Extras/CID-SMILES.gz) and processing it to a small dataset of 50000 SELFIES strings in data/processed/TINY-CID-SELFIES-20, which is already available in the repo.
If you want access to the other datasets (for, say, a larger training run), you can run the following scripts. Warning: you will need plenty of disk space since the uncompressed CID-SMILES is already 8Gb.
download_dataset.pylets you download the dataset and decompress it.smiles_to_strings.pyreads the file atdata/raw/CID-SMILESin chunks and progressively translates the SMILES to SELFIES using The Matter's Lab translator.3 Each processed chunk is appended at the end of a CID-SELFIES file indata/processed/CID-SELFIES.small_selfies.pyfilters all the SELFIES in the dataset that are larger than 20 tokens, outputting a file indata/processed/SMALL-CID-SELFIES-20. Finally, a subset of only 50000 of these 20-length SELFIES is stored indata/processed/TINY-CID-SELFIES-20, which is the file used for training. However, the models and training pipeline are built in such a way that training on the entireSMALL-CID-SELFIES-20should be feasible with a little bit of work.4
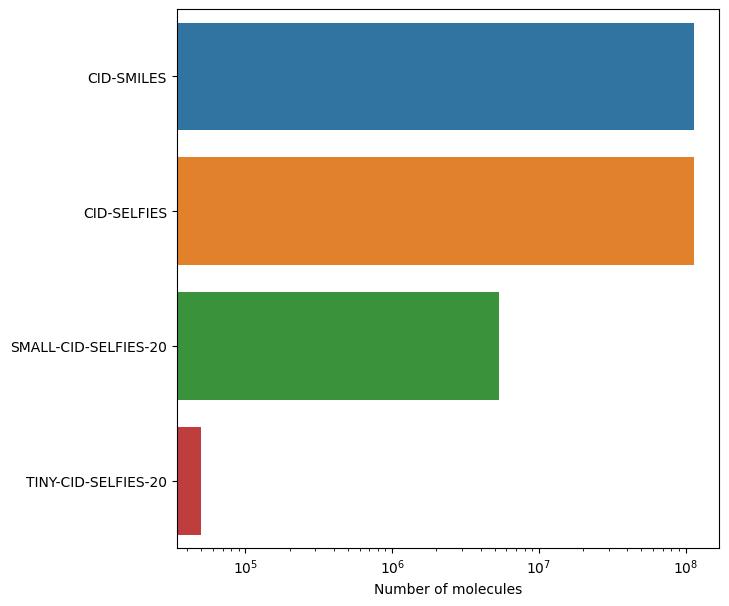
This figure shows the size of these datasets in log-scale.
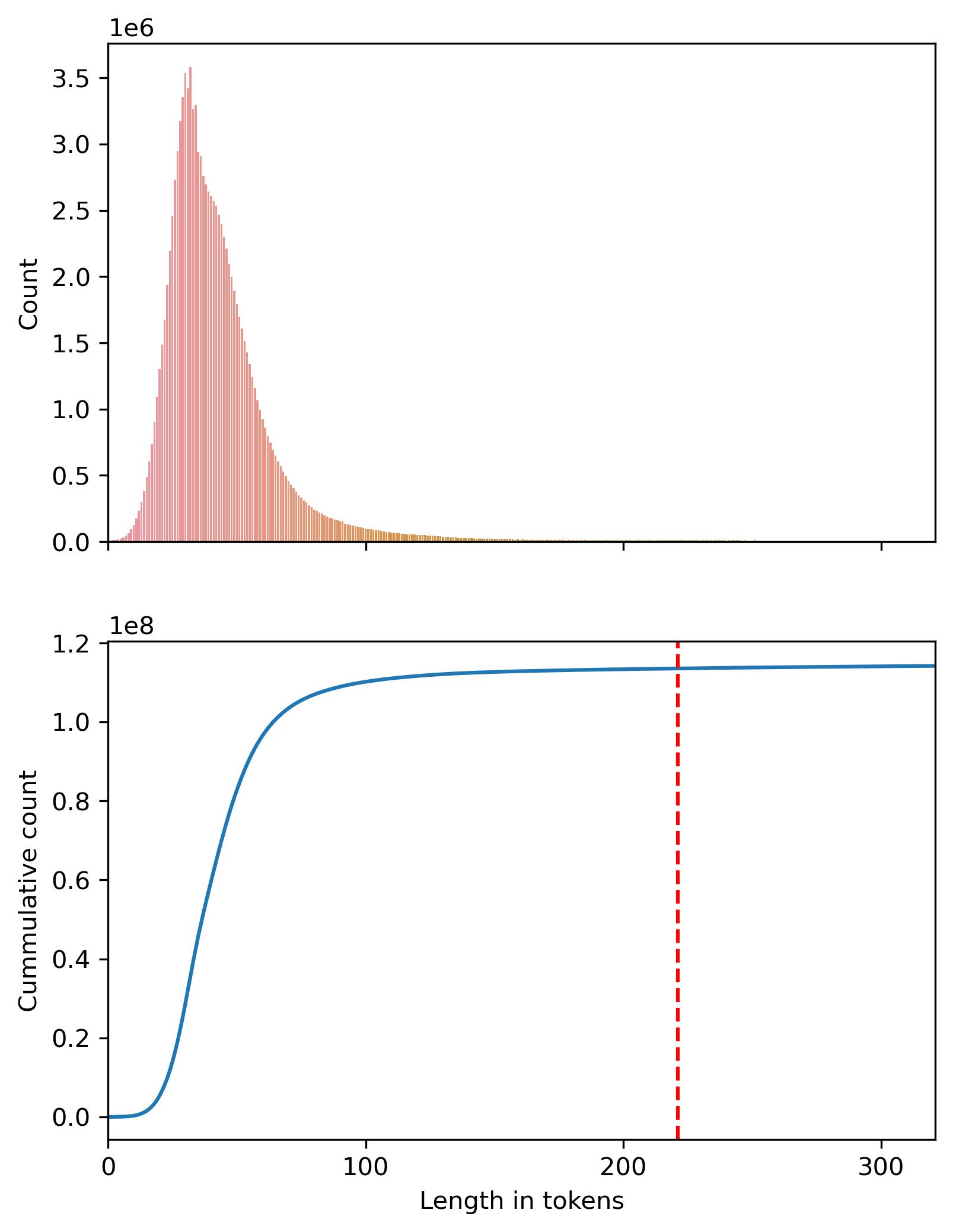
We can also take a look at the distribution of sequence lengths. The following plot shows how most of the mass of token lengths (99%) is concentrated below 221 tokens. We truncate at 320, but the maximum sequence length is 1840.
Ideally, we would also include other datasets like ZINC205, or GuacaMol6. This is included as a TODO at the end of this repo.
The tokenizing is done by hand. The script src/tokenizing/compute_tokens.py takes a database name (like TINY-CID-SELFIES-20) and computes a tokens_dict: Dict[str, int] and saves it inside data/processed/tokens_TINY-CID-SELFIES-20.json. We define the tokens as all the substrings in a SELFIES between square brackets:
# From src/utils/tokens.py
def from_selfie_to_tokens(selfie: str) -> List[str]:
"""
Given a selfie string, returns a list of all
occurences of [.*?] in the string (i.e. whatever
is between square brackes).
"""
return list(re.findall(r"\[.*?\]", selfie))This doesn't scale. Smarter tokenizers are available out there. For example, check MolGen.
We implement a simple MLP Variational Autoencoder.
# Inside the model's definition in src/models/vae.py
# Define the input length: length of a given SELFIES
# (always padded to be {max_length}), times the number of tokens
self.input_length = max_token_length * len(self.tokens_dict)
# Define the model
self.encoder = nn.Sequential(
nn.Linear(self.input_length, 1024),
nn.ReLU(),
nn.Linear(1024, 512),
nn.ReLU(),
nn.Linear(512, 256),
nn.ReLU(),
)
self.encoder_mu = nn.Linear(256, latent_dim)
self.encoder_log_var = nn.Linear(256, latent_dim)
# The decoder, which outputs the logits of the categorical
# distribution over the vocabulary.
self.decoder = nn.Sequential(
nn.Linear(latent_dim, 256),
nn.ReLU(),
nn.Linear(256, 512),
nn.ReLU(),
nn.Linear(512, 1024),
nn.ReLU(),
nn.Linear(1024, self.input_length),
)After running src/training/training_models.py, a trained model is saved in data/trained_models/VAESelfies_TINY-CID-SELFIES-20.pt (download it here).
This model is nowhere close to state-of-the-art. The goal of this code is to have a toy latent space to run experiments in, or to have a starting point for your new SOTA models!
The script in src/exploration/explore_latent_space.py samples randomly from its latent space. Some of those are at the beginning of this repository.
-
Latent space optimization of MolSkill and QED using Evolutionary Strategies. -
Assess the lengths of all molecules inCID-SELFIES - Scalable tokenizing using MolGen's tokenizer.
- Training on all of
SMALL-CID-SELFIES-20. - Latent space optimization of MolSkill and QED using Bayesian Optimization.
- Include other datasets, like GuacaMol or ZINC20.
- Better models, like an autoregressive VAE using LSTMs, or a transformer.
If you find this code useful, feel free to cite it!
@software{Gonzalez-Duque:VAESelfies:2023,
author = {González-Duque, Miguel},
title = {{Minimal implementation of a Variational Autoencoder on SELFIES representations of molecules}},
url = {https://github.com/miguelgondu/minimal_VAE_on_selfies},
version = {0.1},
date = {2023-05-02}
}