NodeStudio is an open-source node-based imaging processing web platform created to facilitate the design, development, and standardization of medical imaging analysis pipelines. It is designed to allow researchers to easily build, design and deploy advanced algorithms and techniques using a visual programming node editor.
NodeStudio was developed to enable researchers to build complex imaging processing pipelines visually using a computation graph. A visual computation graph is a method for the design of complex algorithms without writing a single line of code. Instead of writing code, the user can visually assemble node networks in a cross-platform web application. NodeStudio gives rapid feedback that reflects network changes. It is designed to be simple to use and doesn’t require the knowledge of Python or any standard programming languages. This platform also supports multi-vendor MRI data, the integration of custom Python code, and the integration of third-party packages the deep learning framework DOSMA. As a tool for musculoskeletal MRI analysis, the platform supports several techniques for quantitative MR analysis.
This project is active development. Demo of application is here. Slides for IWAOI conference here.
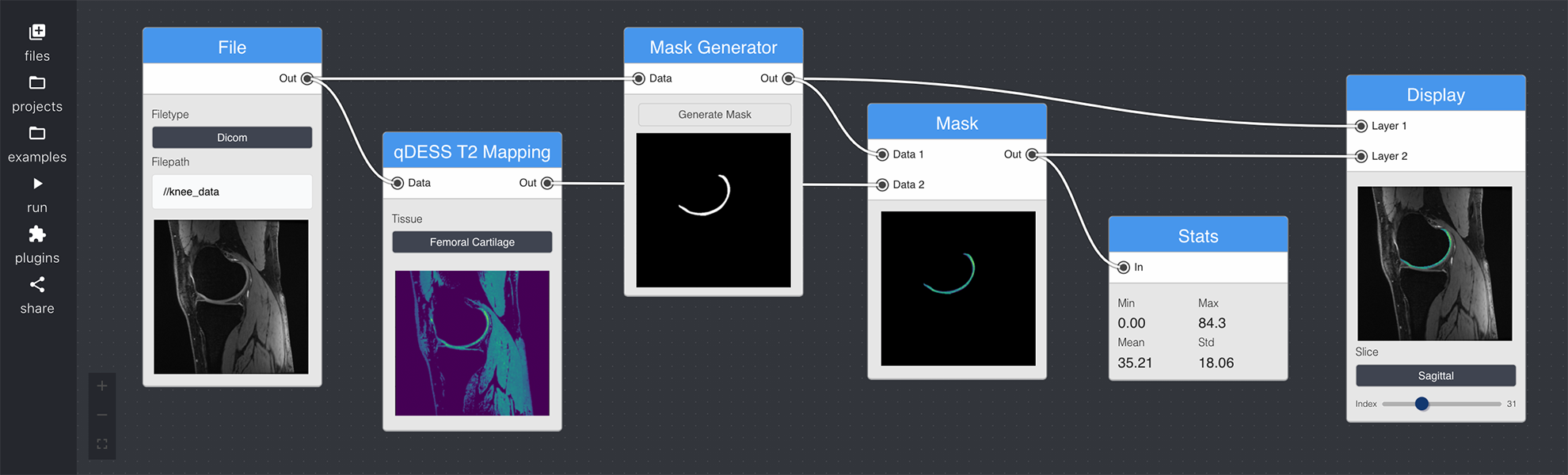 Figure: An Example of a Quantitative MR Analysis Pipeline with NodeStudio User Interface
Figure: An Example of a Quantitative MR Analysis Pipeline with NodeStudio User Interface
Frontend javascript code located in nodestudiojs. Backend python server code located in nodestudio
To run locally frontend: ( From project root directory )
- Install nodejs. (Requires Node 14 or greater)
- Setup nodejs third party packages (uses npm install)
npm run build
- To run frontend react server
npm start
To run locally backend api server: ( From project root directory )
- Setup conda enviroment
You have two options:
- Create a new conda environment from scatch
conda create -n nodestudio python=3.8 conda activate nodestudioThen install packages with pip using requirements file
pip install -r requirements.txtor by individual package
pip install numpy matplotlib scipy scikit-image pip install uvicorn fastapi pydantic pymongo jsonpickle pip install python-socketio websockets gdown tqdm jupyterlab ipywidgets pdf2image pip install pydicom==2.0.0 pymapvbvd dosma tensorflow
- Run to API server
npm run api
Nodestudio is divided into a frontend react application using react and a backend server application using python. The frontend app is located in /nodestudiojs and the Backend app is located in /nodestudio. The frontend application tech stack consists of react with sass. The frontend uses fastapi for a light weight python server.
/nodestudiojs/src
/components - contains react application components
/hooks - contains custom react hooks
/libraries - contains utility functions that don’t have side effects
/models - contains javascript classes used as models in application state
/services - contains functions that touch endpoints & communicate with APIs
/state - contains app state and reducers (functions that modify state & have side-effects.)
/styles - scss styles
/nodestudio
/server.py - server entrypoint
/api - contains fastAPI server code
/controllers - api controllers (should be lightweight)
/routes - api router routes
/graph - contains code for computation graph
/enums - contains NodeType and NodeDetail enums for node designation
/graph - represents computation graph (built from nodes, links and sessions)
/interfaces - interfaces for data types uses pydantic
/link - represents node connections
/node - represents computation node
/nodes - contains list of NodeProps for node properties
/session - represents computation sesssion
/process - contains node processes computation code
/core - base node process i.e. fft, mask, stats
/integration/dosma - dosma intergation
/io - io nodes i.e. file, display
/phantom - phantom for simulation code
/quantiative_maps - quantiative maps for tissue properies i.e. T1, T2
/recon - MR image reconstruction code
/simulation - MR simulation code
/tests - contains unit tests (TODO)
@article{mendoza2022nodestudio,
title = { Nodestudio, A Visual Programming Framework For Musculoskeletal MRI Analysis },
journal = {Osteoarthritis Imaging},
volume = {2},
pages = {100014},
year = {2022},
note = {16th International Workshop on Osteoarthritis ImagingTokyo, JapanJuly 6-8, 2022},
issn = {2772-6541},
doi = {https://doi.org/10.1016/j.ostima.2022.100014},
url = {https://www.sciencedirect.com/science/article/pii/S2772654122000083},
author = {M.A. Mendoza and Z. Huo and Y. Diao and Y. Jin and J. Huang and N.K. Bangerter}
}
In addition using NodeStudio, please also consider citing the work that introduced the method used for analysis.