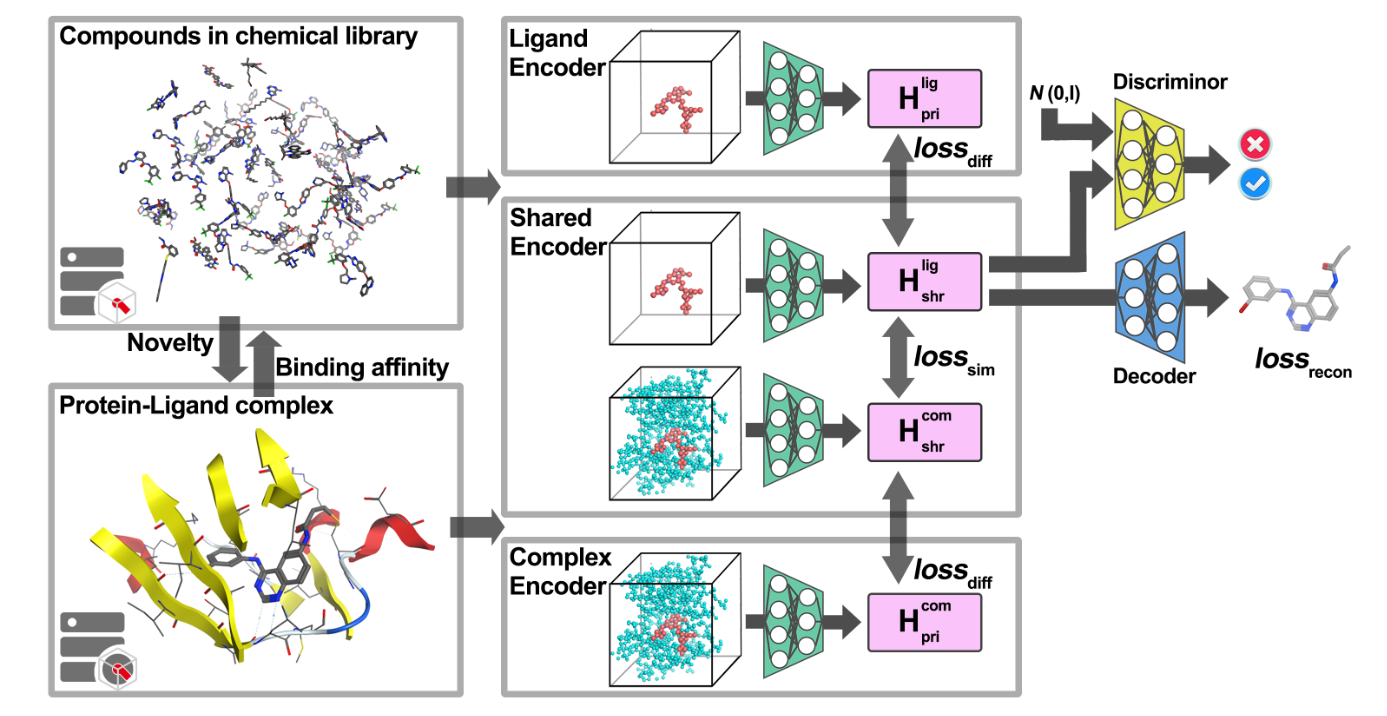
This repository contains the source of RELATION, a software for DL-based de novo drug design.
- Python == 3.7
- pytorch >= 1.1.0
- openbabel == 2.4.1
- RDKit == 2020.09.5
- theano == 1.0.5
- pyscreener README
if utilizing GPU accelerated model training
- CUDA==10.2 & cudnn==7.5
conda env create -f env.yml
To train the RELATION network, the source dataset and target dataset (akt1 and cdk2) must by converted to a 4D-tensor-(19,16,16,16), which means the 3D gird with 19 channels.
python model/data_prepare.py --input ./data/zinc/zinc.csv --output ./data/zinc/zinc.npz --mode 0
python model/data_prepare.py --input ./data/akt1 --output ./data/akt1/akt_pkis.npz --pkidir ./data/akt1.csv --mode 1
Load sourch dataset (./data/zinc/zinc.npz) and target dataset (./data/akt1/akt_pkis.npzor ./data/cdk2/cdk2_pkis.npz).
python model/train.py --epoches 150 --steps 5000 --target akt1 --batchsize 256 --decive 0
Load the ./akt1_relation.pth or ./cdk2_relation.pth generative model, and typing the following codes:
python sample.py --method 0 --numbers 500 --output ./output --target akt1
or you can also use the bayesian optimization in sampling process:
python sample.py --method 1 --numbers 500 --output ./output --target akt1
Or you can use our online RELATION-based server
https://cadd.zju.edu.cn/relation/remode/