Convert simple features to a generic common form that is more general and can be used for a wide variety of data structures.
This is work in progress and at a very early stage. More to come.
library(sf)
#> Linking to GEOS 3.5.1, GDAL 2.2.1, proj.4 4.9.3
## a MULTIPOLYGON layer
nc = st_read(system.file("shape/nc.shp", package="sf"))
#> Reading layer `nc' from data source `/perm_storage/home/mdsumner/R/x86_64-pc-linux-gnu-library/3.4/sf/shape/nc.shp' using driver `ESRI Shapefile'
#> Simple feature collection with 100 features and 14 fields
#> geometry type: MULTIPOLYGON
#> dimension: XY
#> bbox: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
#> epsg (SRID): 4267
#> proj4string: +proj=longlat +datum=NAD27 +no_defsThe common form is the entity tables, objects, paths, vertices and a link table to allow de-duplication of shared vertices. Currently this de-duplication is done on all coordinate fields, but for most applications it will usually be done only in X-Y.
library(scsf)
#> Loading required package: silicate
#>
#> Attaching package: 'scsf'
#> The following objects are masked from 'package:silicate':
#>
#> inlandwaters, minimal_mesh
nc = st_read(system.file("gpkg/nc.gpkg", package="sf"))
#> Reading layer `nc.gpkg' from data source `/perm_storage/home/mdsumner/R/x86_64-pc-linux-gnu-library/3.4/sf/gpkg/nc.gpkg' using driver `GPKG'
#> Simple feature collection with 100 features and 14 fields
#> geometry type: MULTIPOLYGON
#> dimension: XY
#> bbox: xmin: -84.32385 ymin: 33.88199 xmax: -75.45698 ymax: 36.58965
#> epsg (SRID): 4267
#> proj4string: +proj=longlat +datum=NAD27 +no_defs
(bmodel <- PATH(nc))
#> $object
#> # A tibble: 100 x 15
#> AREA PERIMETER CNTY_ CNTY_ID NAME FIPS FIPSNO CRESS_ID BIR74
#> * <dbl> <dbl> <dbl> <dbl> <fctr> <fctr> <dbl> <int> <dbl>
#> 1 0.114 1.442 1825 1825 Ashe 37009 37009 5 1091
#> 2 0.061 1.231 1827 1827 Alleghany 37005 37005 3 487
#> 3 0.143 1.630 1828 1828 Surry 37171 37171 86 3188
#> 4 0.070 2.968 1831 1831 Currituck 37053 37053 27 508
#> 5 0.153 2.206 1832 1832 Northampton 37131 37131 66 1421
#> 6 0.097 1.670 1833 1833 Hertford 37091 37091 46 1452
#> 7 0.062 1.547 1834 1834 Camden 37029 37029 15 286
#> 8 0.091 1.284 1835 1835 Gates 37073 37073 37 420
#> 9 0.118 1.421 1836 1836 Warren 37185 37185 93 968
#> 10 0.124 1.428 1837 1837 Stokes 37169 37169 85 1612
#> # ... with 90 more rows, and 6 more variables: SID74 <dbl>, NWBIR74 <dbl>,
#> # BIR79 <dbl>, SID79 <dbl>, NWBIR79 <dbl>, object <chr>
#>
#> $path
#> # A tibble: 108 x 6
#> ncol type subobject object path ncoords_
#> <int> <chr> <int> <chr> <chr> <int>
#> 1 2 MULTIPOLYGON 1 83cc972d 4ceb1a79 27
#> 2 2 MULTIPOLYGON 1 4e6c51ba 8aa608c2 26
#> 3 2 MULTIPOLYGON 1 220da212 f385bee3 28
#> 4 2 MULTIPOLYGON 1 a9be497e 09d360de 26
#> 5 2 MULTIPOLYGON 2 a9be497e c6b44df2 7
#> 6 2 MULTIPOLYGON 3 a9be497e 12d320f3 5
#> 7 2 MULTIPOLYGON 1 aedde6f5 a9854148 34
#> 8 2 MULTIPOLYGON 1 5205473e 2e295f5f 22
#> 9 2 MULTIPOLYGON 1 e422b229 8bd3fc64 24
#> 10 2 MULTIPOLYGON 1 df2eab80 398f9e7c 17
#> # ... with 98 more rows
#>
#> $vertex
#> # A tibble: 1,255 x 3
#> x_ y_ vertex_
#> <dbl> <dbl> <chr>
#> 1 -81.47276 36.23436 843f3357
#> 2 -81.54084 36.27251 5fdeb201
#> 3 -81.56198 36.27359 1c9f2fab
#> 4 -81.63306 36.34069 1870fd10
#> 5 -81.74107 36.39178 8111a974
#> 6 -81.69828 36.47178 8817bcb8
#> 7 -81.70280 36.51934 23f22fa4
#> 8 -81.67000 36.58965 94b4c37c
#> 9 -81.34530 36.57286 2d3cfd7d
#> 10 -81.34754 36.53791 53c92a50
#> # ... with 1,245 more rows
#>
#> $path_link_vertex
#> # A tibble: 2,529 x 2
#> path vertex_
#> <chr> <chr>
#> 1 4ceb1a79 843f3357
#> 2 4ceb1a79 5fdeb201
#> 3 4ceb1a79 1c9f2fab
#> 4 4ceb1a79 1870fd10
#> 5 4ceb1a79 8111a974
#> 6 4ceb1a79 8817bcb8
#> 7 4ceb1a79 23f22fa4
#> 8 4ceb1a79 94b4c37c
#> 9 4ceb1a79 2d3cfd7d
#> 10 4ceb1a79 53c92a50
#> # ... with 2,519 more rows
#>
#> attr(,"class")
#> [1] "PATH" "sc"
#> attr(,"join_ramp")
#> [1] "object" "path" "path_link_vertex"
#> [4] "vertex"Prove that things work by round-tripping to the PATH model and onto the old fortify approach for ggplot2.
inner_cascade <- function(x) {
tabnames <- silicate:::join_ramp(x)
tab <- x[[tabnames[1]]]
for (ni in tabnames[-1L]) tab <- dplyr::inner_join(tab, x[[ni]])
tab
}
## this just joins everything back together in one big fortify table
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
tab <- bmodel %>% inner_cascade()
#> Joining, by = "object"
#> Joining, by = "path"
#> Joining, by = "vertex_"
library(ggplot2)
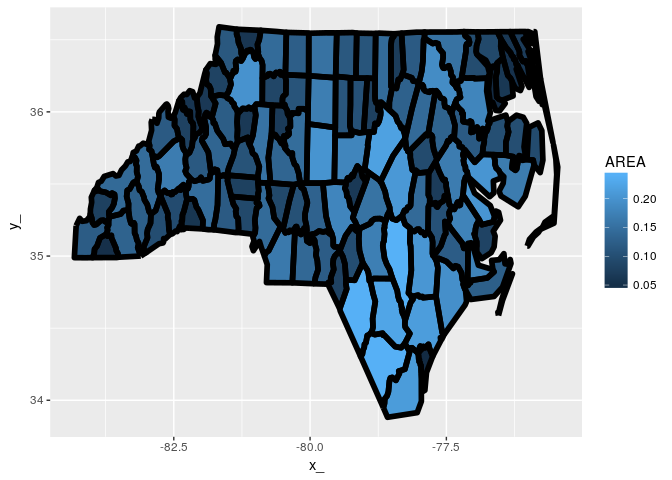
ggplot(tab) + aes(x = x_, y = y_, group = path) +
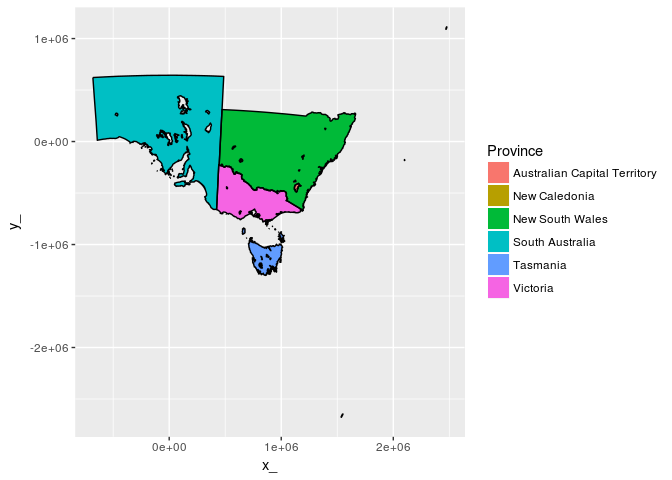
geom_polygon(aes(fill = AREA)) + geom_path(lwd = 2, col = "black") What about polygons with holes and lots of tiny complicated parts.
data("inlandwaters")
iw <- PATH(inlandwaters)
str(iw)
#> List of 4
#> $ object :Classes 'tbl_df', 'tbl' and 'data.frame': 6 obs. of 3 variables:
#> ..$ ID : int [1:6] 103841 103842 103843 103846 103847 103848
#> ..$ Province: chr [1:6] "Australian Capital Territory" "New Caledonia" "New South Wales" "South Australia" ...
#> ..$ object : chr [1:6] "6231a2b3" "0748e369" "4cfb8987" "57130e22" ...
#> ..- attr(*, "sf_column")= chr "geom"
#> ..- attr(*, "agr")= Factor w/ 3 levels "constant","aggregate",..: NA NA
#> .. ..- attr(*, "names")= chr [1:2] "ID" "Province"
#> $ path :Classes 'tbl_df', 'tbl' and 'data.frame': 189 obs. of 6 variables:
#> ..$ ncol : int [1:189] 2 2 2 2 2 2 2 2 2 2 ...
#> ..$ type : chr [1:189] "MULTIPOLYGON" "MULTIPOLYGON" "MULTIPOLYGON" "MULTIPOLYGON" ...
#> ..$ subobject: int [1:189] 1 1 1 1 1 1 1 1 1 1 ...
#> ..$ object : chr [1:189] "6231a2b3" "0748e369" "4cfb8987" "4cfb8987" ...
#> ..$ path : chr [1:189] "e3d22cca" "062fe863" "9811579f" "4a8545d7" ...
#> ..$ ncoords_ : int [1:189] 280 27 7310 68 280 88 162 119 51 71 ...
#> $ vertex :Classes 'tbl_df', 'tbl' and 'data.frame': 30835 obs. of 3 variables:
#> ..$ x_ : num [1:30835] 1116371 1117093 1117172 1117741 1117629 ...
#> ..$ y_ : num [1:30835] -458419 -457111 -456893 -456561 -455510 ...
#> ..$ vertex_: chr [1:30835] "be17eacc" "f45a7f3b" "61b69e20" "87cc6dc7" ...
#> $ path_link_vertex:Classes 'tbl_df', 'tbl' and 'data.frame': 33644 obs. of 2 variables:
#> ..$ path : chr [1:33644] "e3d22cca" "e3d22cca" "e3d22cca" "e3d22cca" ...
#> ..$ vertex_: chr [1:33644] "be17eacc" "f45a7f3b" "61b69e20" "87cc6dc7" ...
#> - attr(*, "class")= chr [1:2] "PATH" "sc"
#> - attr(*, "join_ramp")= chr [1:4] "object" "path" "path_link_vertex" "vertex"
tab <- iw %>% inner_cascade()
#> Joining, by = "object"
#> Joining, by = "path"
#> Joining, by = "vertex_"
library(ggplot2)
ggplot(tab) + aes(x = x_, y = y_, group = path) +
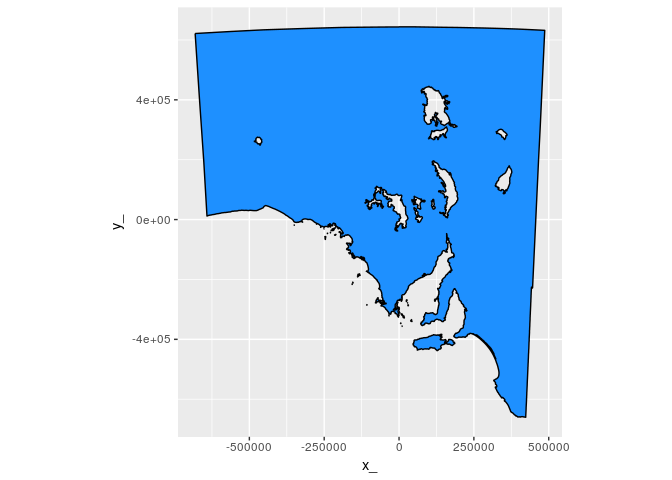
ggpolypath::geom_polypath(aes(fill = Province)) + geom_path(col = "black") ggplot(tab %>% filter(Province == "South Australia")) + aes(x = x_, y = y_, group = path) +
ggpolypath::geom_polypath(fill = "dodgerblue") + geom_path(col = "black") + coord_fixed()