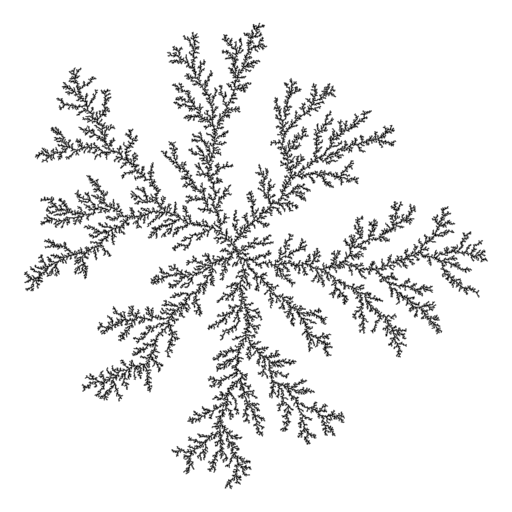
An arbitrary-dimensional diffusion-limited aggregation simulator.
dla-nd supports the following features
- Automatic, persistent, adaptive 2^D-tree space subdivision
- Statistically-correct particle motion bias (bulk motion) numerically solves quartic equation each step
- Stubbornness (Nittman and Stanley's artificial surface tension model)
- Tunable stickiness and grippiness
- 2-d or 3-d density field construction
- Raw, Radiance, and generic OBJ output for particle positions and connectivities
- PNG and ASCII PGM output for particle locations and density field
- Restart from an old simulation, or merge multiple simulations using raw .part files
See some images and more at the old homepage.
This software uses the CMake build system, which is available on Windows, OSX, and Linux. The only external dependency is libpng, which is usually installed on most systems. If not, you can get both dependencies with (Red Hat/Fedora, Debian/Ubuntu, OSX/brew):
sudo dnf install cmake libpng-devel
sudo apt-get install cmake libpng-dev
brew install cmake libpng
Once that's settled, from the directory with this README, run:
mkdir build
cd build
cmake -DCMAKE_BUILD_TYPE=Release ..
make
and several binary executables will be made. If you want to build a specific executable for 1D through 5D, just ask for it directly:
make dla-3d
You no longer need to use an input file to run dla-nd, but I haven't documented the options well. See the setup.c file for those. But to get started with some fun stuff, try:
dla-2d -p 0 0 -every 1000 -end 100000 -zone -2 2 -2 2 -dens -res 1000
This will run a 2D sim which starts with a single particle at 0,0, writes a 1000x1000 pixel PNG image out every 1000 steps, in which the particles are rendered onto a density field that spans -2..2 in x and y.
dla-3d -p 0 0 0 -every 10000 -end 100000 -zone -1 1 -1 1 -1 1 -slices -res 200 -seg
The above command will perform a similar job, but will write 200 PNG files every 10000 steps (slices) and also write an OBJ-like "seg" file containing the vector geometry. This parts0001.seg file can be manipulated with my stickkit program.
Windows users can also run "cmd" to enter the DOS-like command prompt. From there, you can run any of the commands above.
- Speed up by blocking: instead of linked lists of particles, put them in blocks of data per tree node
- Speed up by multi-threading: allow many independent traversals, but lock when modifying the tree
- Start using command-line options instead of an input file
- Fill this out with descriptions of
-gripand-stickand bulk motion, etc. Maybe some images, too.
I don't get paid for writing or maintaining this, so if you find this tool useful or mention it in your writing, please please cite it by using the following BibTeX entry.
@Misc{dland2014,
author = {Mark J.~Stock},
title = {dla-nd: An arbitrary-dimensional diffusion-limited aggregation simulator},
howpublished = {\url{https://github.com/markstock/dla-nd}},
year = {2014}
}