This is the official PyTorch implementation of the paper: Weakly Supervised Learning of Cortical Surface Reconstruction from Segmentations (MICCAI 2024). We proposed CoSeg, a deep learning-based cortical surface reconstruction framework weakly supervised by pseudo ground truth brain segmentations.
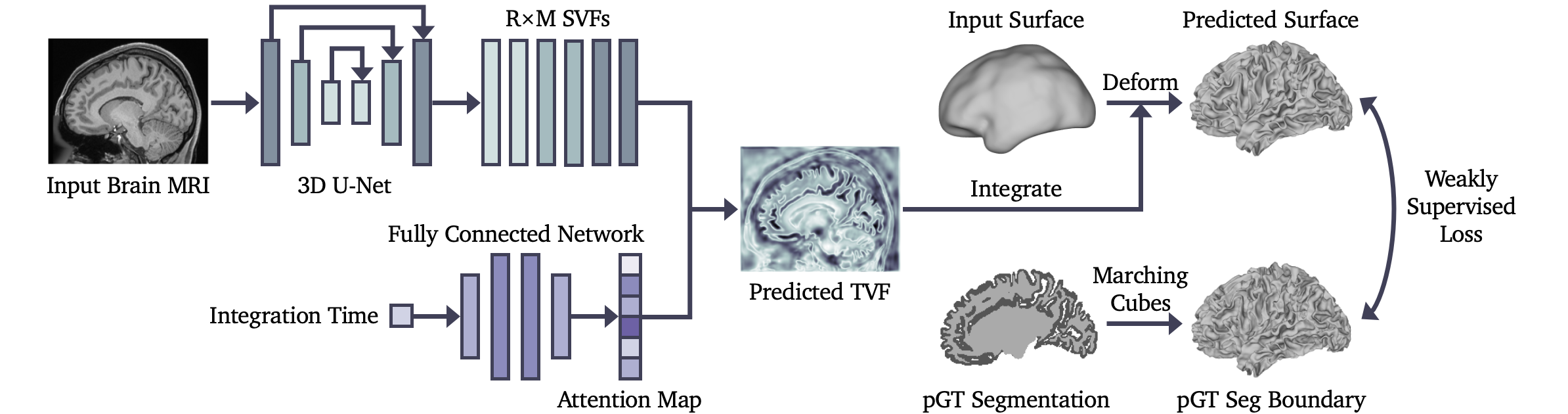 The architecture of the Temporal Attention Network (TA-Net).
The architecture of the Temporal Attention Network (TA-Net).
 (a) The cGM segmentation boundary; (b) The expected pial surface; (c) Bidirectional Chamfer distance between the cGM segmentation boundary and input white surface; (d) The boundary loss, i.e., single-directional Chamfer distance; (e) The inflation loss between the vertex displacement and the normal vector of the input white surface.
(a) The cGM segmentation boundary; (b) The expected pial surface; (c) Bidirectional Chamfer distance between the cGM segmentation boundary and input white surface; (d) The boundary loss, i.e., single-directional Chamfer distance; (e) The inflation loss between the vertex displacement and the normal vector of the input white surface.
Our CoSeg framework requires the following dependencies:
- PyTorch == 1.7.1
- PyTorch3D == 0.4.0 (for training only)
- Nibabel == 3.2.1
- Trimesh == 3.9.15
- NumPy == 1.23.5
- SciPy == 1.10.0
- Scikit-image == 0.18.1
- ANTsPy == 0.3.4 (for registration only)
This paper is evaluated on the HCP young adult dataset and the dHCP fetal dataset. For the HCP data, we use the T1w brain MRI T1_restore.nii.gz and cortical ribbon segmentation ribbon.nii.gz under the directory ./MNINonLinear of each subject for training. The T1w brain images and segmentations have already been affinely aligned to the MNI-152 space. To preprocess the HCP data, please run:
python ./data/data_preprocess_hcp.py --orig_dir='/YOUR_HCP_DATA/'
The preprocessed data will be saved to ./data/hcp/.
For dHCP fetal data, we use BOUNTI fetal brain MRI segmentation pipeline to generate pseudo ground truth brain tissue segmentation masks. To preprocess the dHCP data, please run:
python ./data/data_preprocess_dhcp.py --orig_dir='/YOUR_DHCP_DATA/'
The preprocessed data will be saved to ./data/dhcp/. The T2w fetal brain MR images are affinely aligned to a 36-week dHCP neonatal atlas ./template/dhcp_fetal_week36_t2w.nii.gz. The labels of BOUNTI brain tissue segmentations are merged and split into left and right brain hemispheres to create cortical ribbon segmentations for training.
To train CoSeg for left/right white/pial surface reconstruction on the HCP/dHCP dataset, please run:
python train.py --data_type='dhcp'\
--surf_type='pial'\
--surf_hemi='left'\
--tag='EXP_ID'\
--w_edge=0.5\
--w_nc=5.0\
--w_inflate=5.0\
--n_epoch=200\
--device='cuda:0'
where data_type=['hcp','dhcp'] is the name of the dataset, surf_type=['white','pial'] is the type of the surface, surf_hemi=['left','right'] is the brain hemisphere, tag is a string to identify your experiments, and n_epoch is the number of epochs for training. w_edge, w_nc, and w_inflate are the weights for the edge length loss, normal consistency loss, and inflation loss.
The training process loads data from ./data/data_type/ and saves the model checkpoints to the ./ckpts/data_type/ directory. Please note that the training of white surface reconstruction is a prerequisite for the training of pial surface reconstruction. For more details of all arguments please run python train.py --help.
For the inference, please run:
python pred.py --data_dir='/YOUR_DATASET/YOUR_MRI.nii.gz'\
--save_dir='/YOUR_RESULT/'\
--data_type='dhcp'\
--surf_hemi='left'
where data_dir is the file name of the preprocessed brain MRI. The predicted surfaces will be saved to save_dir in GIfTI format .surf.gii.