|

|
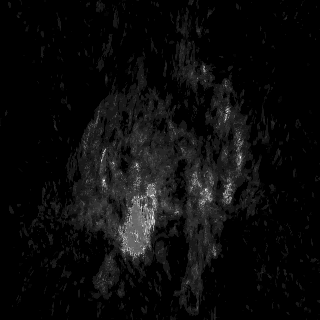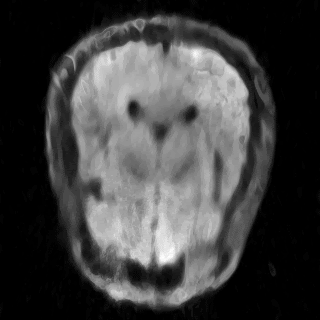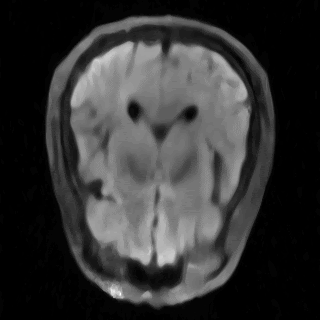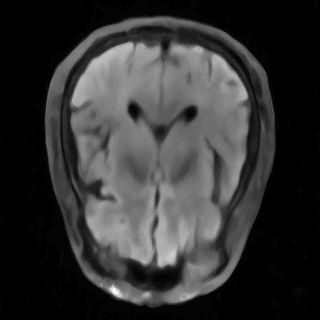
This is the repository for the project "Learning Neural Implicit Representations of MRI Data" of the ADLM Praktikum, supervised by Wenqi Huang and Robert Graf. Within this repository, our objective is to delve into and compare the most recent advancements in Implicit Neural Representations (INRs) for accurately modeling MRI data. The primary goal is to assess and compare the effectiveness of various methods for processing MRI data, both in image space and k-space. Our particular focus is on multi-coil data. The assets folder contains some results.
We use FastMRI's multi coil dataset for our experiments. For ease of use with local config files, download the data and symlink it to ./data in the project's root directory.
conda create --name inr
conda activate inr
pip install -r requirements.txt
To run an experiment create a configuration file as in here. Some basic configuration files already exist and the basic usage is
python src/train.py --config path/to/config
To run the experiments with multiple samples use the data samples config files:
python src/train.py --config path/to/config --data_samples path/to/data_samples
To train a multiscale network on k-space data, run the following command:
python src/train_kspace_multiscale.py --config path/to/config
Our Multi-Scale Network is based on a light-weight pre-processing step that clusters the available k-space data. Note that the clustering setup must be included in the config as in this file.
To run the experiments with multiple samples use the data samples config files:
python src/train_kspace_multiscale.py --config path/to/config --data_samples path/to/data_samples
The configuration file is the main entry point to modify the behavior of the INR during training.
-
Single-Scale:
["SIREN", "WIRE", "WIRE2D", "FFN", "FOURIER", "GABOR"] -
Multi-Scale:
["Fourier", "BoundedFourier"]
In particular, FourierNet and GaborNet are presented in [3]. We base our Multi-scale networks off of [4].
Available Criteria: ["L2", "MSLE", "FFL", "L1", "HDR", "tanh"]
In Particular, HDR loss is presented in [1] and Tanh loss in [2]. LSL is a lightweight HDR loss.
Generally, HDR is superior in recovering the K-space periphery if set up correctly. Otherwise, L2 and Tanh are good choices.
Available Normalizations: ["abs_max", "max" "max_std", "coil"]
Generally, per-coil normalization "coil" and normalizing by the absolute max "max" are good choices.
Here we provide an example on how to set up the ring clustering
partition:
no_steps: 40 # Initial number of rings
no_models: 4 # Final number of rings after clustering
Here we provide an example on how to set up the undersampling param
# Supported formats:
# grid-x*y example: grid-3*3
# random_line-p example: random_line-0.5
# radial-acc_fac example: radial-4
# none No undersamping
undersampling: grid-2*1
Note: Line sampling has not been implemented separately; instead, it utilizes our grid sampling code. So:
For Column sampling use grid-n*1 where n is the accelartion in column axis
For Row sampling use grid-1*n where n is the accelartion in row axis
To set the dataloader to load batches per coil, please set the parameter per_coil: True in the config file.
If per coil batching is used, one can also set use_tv to calculate the total variation loss on the coil to enforce spatial smoothness as in [5].
To perform a hyperparameter search, one can use two methods - Grid Search and Random Search. To use a method use a sample for model WIRE as below:
{
"method": "random",
"num_search": 2,
"search_space": {
"batch_size":{
"values": [10000, 25000],
"type": "int"
},
"lr": {
"values": [0.00001, 0.00003],
"type": "log"
},
"net.network_depth": {
"values": [2, 4],
"type": "int"
},
"net.scale": {
"values": [10, 20],
"type": "int"
},
"normalization": {
"values": ["coil", "gaussian_blur", "max_std", "max"],
"type": "item"
}
}
}
The hyperparameters must be added in the key search_space and each hyperparameter must have a type belonging to ['log', 'int', 'float', 'item'].
A nested hyperparameter like scale should be appended after a '.' with it's parent key, like net.scale.
To perform the search use the following command:
python src/hp_search_script.py --config path/to/config --hp_config path/to/hp_config
Moreover, we have also added a series of unit tests. These tests can be found under src/test folder, inluding tests for the dataloader, regularization and undersampling.
Note: Dataloader's unit tests require (.h5) data to work. Please download that file and modify the unit test accordingly.
[1] Huang, W., Li, H., Cruz, G., Pan, J., Rueckert, D., & Hammernik, K. (2022). Neural Implicit k-Space for Binning-free Non-Cartesian Cardiac MR Imaging. Information Processing in Medical Imaging.
[2] Chen, X., Liu, Y., Zhang, Z., Qiao, Y., & Dong, C. (2021). HDRUNet: Single Image HDR Reconstruction with Denoising and Dequantization. 2021 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), 354-363.
[3] Fathony, R., Sahu, A., Willmott, D., & Kolter, J.Z. (2021). Multiplicative Filter Networks. International Conference on Learning Representations.
[4] Lindell, D.B., Veen, D.V., Park, J.J., & Wetzstein, G. (2021). Bacon: Band-limited Coordinate Networks for Multiscale Scene Representation. 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), 16231-16241.
[5] Block, K. T., Uecker, M., & Frahm, J. (2007). Undersampled radial MRI with multiple coils. Iterative image reconstruction using a total variation constraint. Magnetic resonance in medicine, 57(6), 1086–1098. https://doi.org/10.1002/mrm.21236