SEREEGA is a modular, general-purpose open-source MATLAB-based toolbox to generate simulated toy data mimicking event-related electroencephalography (EEG) data.
Reference:
(See also this PDF mirror, or this bioRxiv preprint.)
- Introduction
- Tutorial
- Installation
- Configuration
- Obtaining a lead field
- Picking a source location
- Orienting a source dipole
- Defining a source activation signal
- Components and scalp data
- Finalising a dataset
- Variability
- Noise and multiple signal classes
- A note on components and sources
- More signal types
- Random class generation and multiple components
- Source identification
- Component variability
- Relativity of the lead field units
- Using the GUI
- Extending SEREEGA
- Sample code
- Contact
- Special thanks and acknowledgements
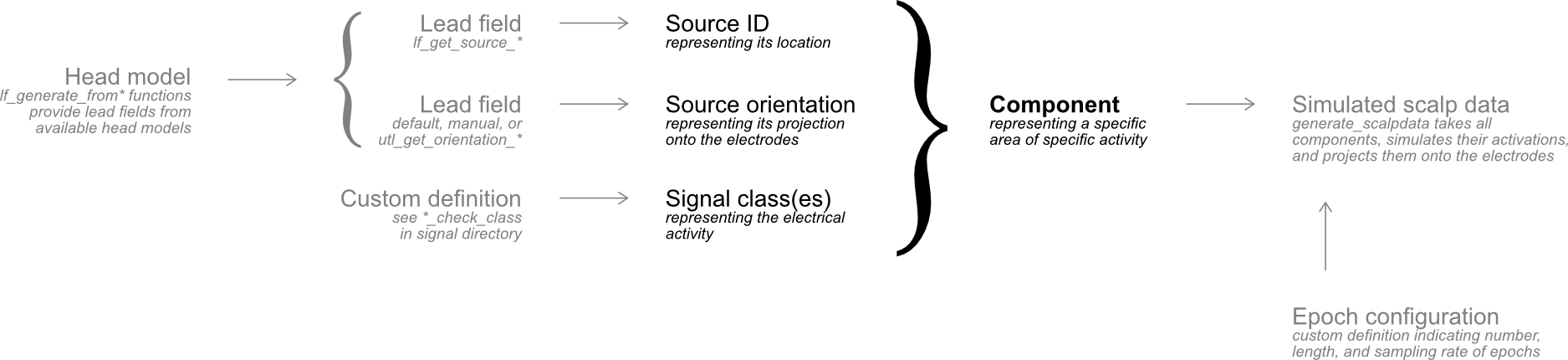
SEREEGA is an open-source MATLAB-based toolbox to generate simulated, event-related EEG toy data. Starting with a forward model obtained from a head model or pre-generated lead field, dipolar components can be defined. Each component has a specified position and orientation in the head model. Different activation signals can be assigned to these components. EEG data is simulated by projecting all activation signals from all components onto the scalp and summing them together.
SEREEGA is modular in that different head models and lead fields are supported, as well as different activation signals. Currently, SEREEGA supports the New York Head (ICBM-NY) pre-generated lead field, the Pediatric Head Atlases pre-generated lead fields, the HArtMuT (Head Artefact Model using Tripoles) pre-generated lead fields, and FieldTrip custom lead field generation. Four types of activation signals are provided: ERP (event-related potential, i.e. systematic deflections in the time domain), ERSP (event-related spectral perturbation, i.e. systematic modulations of oscillatory activations), noise (stochastic processes in the time domain), and signals based on an autoregressive model. A final data class allows the inclusion of any already existing time series as an activation signal.
SEREEGA is intended as a tool to generate data with a known ground truth in order to evaluate neuroscientific and signal processing methods, e.g. blind source separation, source localisation, connectivity measures, brain-computer interface classifier accuracy, derivative EEG measures, et cetera.
As an example, the following code produces an EEGLAB dataset containing 100 epochs of 64-channel EEG data, each consisting of the summed activity of 64 simulated sources spread across the brain. The final dataset includes a ground-truth ICA decomposition that can be used to verify the accuracy of newly calculated decompositions.
% configuring for 100 epochs of 1000 ms at 1000 Hz
config = struct('n', 100, 'srate', 1000, 'length', 1000);
% obtaining a 64-channel lead field from ICBM-NY
leadfield = lf_generate_fromnyhead('montage', 'S64');
% obtaining 64 random source locations, at least 2.5 cm apart
sourcelocs = lf_get_source_spaced(leadfield, 64, 25);
% each source will get the same type of activation: brown coloured noise
signal = struct('type', 'noise', 'color', 'brown', 'amplitude', 1);
% packing source locations and activations together into components
components = utl_create_component(sourcelocs, signal, leadfield);
% simulating data
data = generate_scalpdata(components, leadfield, config);
% converting to EEGLAB dataset format, adding ICA weights
EEG = utl_create_eeglabdataset(data, config, leadfield);
EEG = utl_add_icaweights_toeeglabdataset(EEG, components, leadfield);Below is a brief tutorial running through the basics of SEREEGA. This tutorial focuses on scripting, i.e. assigning values to variables and calling SEREEGA's functions to simulate data. The GUI is only discussed later, as the scripts provide a more thorough understanding of the toolbox, and the GUI is based on these scripts.
Note that the code examples are more or less intended to follow each other. Separate pieces of code may not work without having executed the code in earlier sections.
For a detailed overview of all functions and their capabilities, please look at the documentation in the individual scripts themselves.
Download SEREEGA to your computer and add all its (sub)directories to MATLAB's path.
It is recommended to have MATLAB 2014b or higher with the DSP and Parallel Computing toolboxes installed. SEREEGA's core functionality requires R2013b. This is primarily due to the use of addParameter rather than addParamValue; if your MATLAB does not support addParameter, try exchanging all those references with addParamValue. That should restore basic functionality. Some plotting functions rely on the boundary function introduced in R2014b. Some signal generation functions depend on the DSP toolbox version 8.6 (R2014a) or higher. Data simulation tries to use parallel processing from the Parallel Computing Toolbox where available.
EEGLAB is used for a number of functions, and should be started before generating a lead field as EEGLAB's readlocs is used to add channel location information. SEREEGA was tested with EEGLAB 13.6.5b and 2021.1.
When using the New York Head, as in this tutorial, make sure you have the New York Head (ICBM-NY) lead field in MATLAB format in your path. Similarly, the Pediatric Head Atlases and HArtMuT head models must also first be obtained separately, if you intend to use those. When using FieldTrip to generate a custom lead field, the file /fileio/ft_read_sens from the FieldTrip directory will be necessary. FieldTrip can be installed as an EEGLAB plug-in. To obtain a lead field from Brainstorm, Brainstorm needs to be in MATLAB's path. When some functionality in SEREEGA relies on external files which you don't have, a warning will appear with instructions on where to find them. Reasons for these files not having been included out of the box include file size and license conflicts.
Make sure that EEGLAB has been started for this tutorial. We will be using the lead field from the New York Head, thus, the file sa_nyhead.mat should be in the path, available here.
The general configuration of the to-be-simulated epochs is stored in a structure array. The following fields are required.
epochs = struct();
epochs.n = 100; % the number of epochs to simulate
epochs.srate = 1000; % their sampling rate in Hz
epochs.length = 1000; % their length in msAdditional, general parameters can be added here as well, such as prestim to indicate a pre-stimulus time period (in ms) and shift the x-axis accordingly, or marker to indicate the event marker accompanying each epoch at t=0.
The first step in SEREEGA is to obtain a lead field. We can do this by either generating one using FieldTrip, or by using (a part of) a pre-generated lead field. The lead field determines the head model and the electrodes (channels) for the simulation.
We can indicate the electrode locations we want to use by indicating their channel labels, or by indicating a predefined electrode montage. For example, to simulate a predefined 64-electrode montage using the ICBM-NY pre-generated lead field:
lf = lf_generate_fromnyhead('montage', 'S64');S64 refers to a selection, i.e. a montage, of 64 channels from the 10-20 system. The file utl_get_montage contains all available montages. If the channel montage you want is not available there, you can either add it to that file, or indicate the individual channel labels when obtaining the lead field. For example, to only simulate the centre line, we can use:
lf2 = lf_generate_fromnyhead('labels', {'Fz', 'Cz', 'Pz', 'Oz'})When not indicating any montage or labels, most functions will default to using all available electrode positions.
When generating the lead field using FieldTrip, we can also indicate the resolution of the source model (i.e. the density of the source grid):
lf2 = lf_generate_fromfieldtrip('montage', 'S64', 'resolution', 5);We can inspect the channel locations in the obtained lead field and their relation to the brain sources by plotting them:
plot_headmodel(lf);
plot_headmodel(lf, 'style', 'boundary', 'labels', 0);Most lead fields supported by SEREEGA are more or less pre-packaged. A separate software called Brainstorm allows you to create personalised lead fields (aka headmodels in Brainstorm) based on MRI scans. The lf_generate_frombrainstorm enables you to convert a lead field generated in Brainstorm to a format usable by SEREEGA.
To use this function, Brainstorm needs to be installed and in MATLAB's path.
To use your own, personalised lead fields, you need to provide three file paths (plus one optional) to the following files generated by Brainstorm:
- The headmodel mat file generated in Brainstorm.
- The channel location mat file used for the generation of the headmodel in Brainstorm.
- The structural MRI mat file to which the electrodes have been coregistered.
- (Optional) the atlas file if used.
A description of how to obtain these files is included in the help section of lf_generate_frombrainstorm. The command then becomes:
% use personal files
lf = lf_generate_frombrainstorm( ...
'headmodel', 'PATH_TO\my_headmodel.mat', ...
'chanloc', 'PATH_TO\my_chanloc.mat', ...
't1', 'PATH_TO\my_T1_MRI.mat', ...
'atlas', 'PATH_TO\my_atlas.mat');The function also comes with a precomputed head model and associated files, which it uses by default if no other files are supplied, i.e., when you run:
% use defaults
lf = lf_generate_frombrainstorm();Note that the atlas remains optional. Therefore, to use this, you would run:
lf = lf_generate_frombrainstorm('atlas', 'scout_Mindboggle_62.mat');These default files were obtained as follows:
- EGI HydroCel 256 channel locations already coregistered by Brainstorm to the default T1 image (next point)
- ICBM152 T1 image used in the default anatomy by Brainstorm
- Surface Headmodel generated from the above files by applying OpenMEEG to the BEM surfaces and costraining the dipoles to the cortex surface
- Mindboggle 62 atlas
Brainstorm headmodels are expressed in lf_generate_frombrainstorm automatically converts these into
The conversion provides more realistic units, which helps when defining and interpreting results, especially for ERPs. However, if you desire to work with the International System, you can set the scaleUnits argument to 0 to turn the conversion off. The help file of the function provides more details about this.
Furthermore, note that this function converts the coordinate system from the Subject-Coordinate-System (SCS) used in Brainstorm, to the MNI system used in SEREEGA. You can turn this option off by setting the parameter useMNI to 0.
Finally, as Brainstorm uses the SCS, the channel and dipole coordinates are expressed in meters. lf_generate_frombrainstorm converts these measures to mm, for consistency with the other leadfields. The argument useMm can be used to override this.
The lead field contains a number of 'sources' in the virtual brain, i.e. possible dipole locations plus their projection patterns to the selected electrodes. In order to simulate a signal coming from a given source location, we must first select that source in the lead field.
We can get a random source, and inspect its location:
source = lf_get_source_random(lf);
plot_source_location(source, lf);Or, if we know from where in the brain we wish to simulate activity, we can get the source from the lead field that is nearest to that location's coordinates, for example, somewhere in the right visual cortex:
source = lf_get_source_nearest(lf, [20 -75 0]);
plot_source_location(source, lf, 'mode', '3d');Other options to obtain source locations are lf_get_source_inradius to get all sources in a specific radius from either given location coordinates, or from another source in the lead field. If you want more than one source picked at random but with a specific spacing between them, use lf_get_source_spaced.
Starting with SEREEGA v1.4.0, lead fields can have an associated atlas. This means that each individual source in the lead field can be associated with a specific region of the brain. Depending on the lead field and atlas used, some sources may be in the 'Prefrontal cortex', others in the 'Superior temporal gyrus', and others in 'Brodmann area 32', for example. This allows you to pick a source based on the names of these regions, as opposed to using numerical coordinates.
As of December 2022, the New York Head also comes with such an atlas. (If you have downloaded this head model before that time, you may want to download it again.) The following code uses plot_headmodel to visualise the region taken up by one particular region, defined as the right insular cortex. It then uses lf_get_source_random we saw before to obtain a random source from within this region.
plot_headmodel(lf, 'region', {'Brain_Right_Insular_Cortex'}, 'labels', 0);
source_insularrnd = lf_get_source_random(lf, 'region', {'Brain_Right_Insular_Cortex'});Other functions allow you to e.g. obtain a source closest to some calculated 'middle' of a region. All lf_get_source_* functions can be constrained to specific regions.
source_insularmid = lf_get_source_middle(lf, 'region', {'Brain_Right_Insular_Cortex'});
plot_source_location(source_insularmid, lf, 'mode', '3d');Note that few lead fields come with an appropriate atlas. The latest version of the New York Head does, and the HArtMuT head model has a detailed atlas for all its non-brain sources, using those obtained from the New York Head for its cortical sources. Currently, no other supported third-party lead field comes with an atlas. A utility called mni2atlas has been made compatible with SEREEGA through the utl_add_atlas_frommni2atlas function, allowing source coordinates from a lead field to be mapped according to a number of available atlases. However, this may result in a poorly-fitting solution, so use with caution.
The sources in the lead field are represented by dipoles at specific locations. These dipoles can be oriented in space into any direction: they can be pointed towards the nose, or the left ear, or anywhere else. Their orientation is indicated using an [x, y, z] orientation array. Along with its location, a dipole's orientation determines how it projects onto the scalp.
Our source's projections onto scalp along the X (left-to-right), Y (back-to-front), and Z (bottom-to-top) directions, for example, look like this:
plot_source_projection(source, lf, 'orientation', [1 0 0], 'orientedonly', 1);
plot_source_projection(source, lf, 'orientation', [0 1 0], 'orientedonly', 1);
plot_source_projection(source, lf, 'orientation', [0 0 1], 'orientedonly', 1);These projections can be linearly combined to get any simulated dipole orientation. In the ICBM-NY lead field, default orientations are included, which orient the dipole perpendicular to the cortical surface. If no orientation is provided, this default orientation is used. FieldTrip-generated lead fields and the Pediatric Head Atlases have no meaningful default orientation and thus revert to all-zero orientations. It is thus necessary to explicitly indicate an orientation for these lead fields to work. For that, you can use utl_get_orientation_random to get a random orientation, utl_get_orientation_pseudoperpendicular for an orientation pointing outward toward the scalp surface, or utl_get_orientation_pseudotangential for an orientation that attempts to approximate a tangent to the surface.
plot_source_projection(source, lf, 'orientation', [1, 1, 0]);
plot_source_projection(source, lf);When looking at source localisation results in EEG literature, authors usually report the location of a source, but not its orientation per se. What they do report, however, is its projection pattern, usually in the form of topoplots like the ones we plotted in this section. To mimic known effects, you could thus either find a souce from your region of interest whose default orientation results in the projection pattern you are seeking to mimic, or you can pick a source and adjust its orientation to match the desired projection.
Note that the plot_source_projection function used here merely plots the projection pattern at the indicated orientation; it does not by itself change the orientation of any source in the simulation. The actual source orientations to be used in the simulation are indicated at the level of components, described further below.
We now have a source's location and its orientation, i.e., we now know exactly where in the brain this source is, and how it projects onto the scalp. Next, we must determine what, actually, is to be projected onto the scalp. That is, we must define the source's activation pattern.
Let us consider an event-related potential (ERP). In SEREEGA, an ERP activation signal is defined by the latency, width, and amplitude of its peak(s). We store activation definitions in classes, in the form of structure arrays:
erp = struct();
erp.peakLatency = 500; % in ms, starting at the start of the epoch
erp.peakWidth = 200; % in ms
erp.peakAmplitude = 1; % in microvoltThe above thus describes an ERP where a single positive peak, 200 ms wide, is centred at the 500 ms mark, where its peak amplitude is +1 microvolt.
(Note that the amplitude indicated here is the amplitude as it is generated at the source; the final simulated amplitude at the scalp depends on the lead field, and on possibly interfering other signals.)
This toolbox actually works with more than these three parameters for ERP class definitions. Those other parameters are optional and we will look at them later. They are, however, required for the further functioning of this toolbox. To make sure the class is properly defined, including all currently missing parameters, we can use the following function and see the newly added values. (Alternatively, we could indicate all parameters by hand.)
erp = utl_check_class(erp, 'type', 'erp')(You may notice one of the parameters of the final class is its type, which is set to erp. This is how SEREEGA knows how to interpret this class. If we had manually added this field to our class definition, we no longer would have needed to pass this information to utl_check_class, and instead could have simple called erp = utl_check_class(erp).)
We can now plot what this ERP would look like. For this, we also need to know the length of the epoch and the sampling rate, which we defined earlier in our epochs configuration struct.
plot_signal_fromclass(erp, epochs);An ERP activation class can have any number of peaks. For n peaks, you should then also indicate n latencies, n widths, n amplitudes, et cetera--one for each peak. For example, erp = struct('peakLatency', [450, 500], 'peakWidth', [200, 200], 'peakAmplitude', [-1, 1]) produces an ERP with two peaks.
Having defined both a signal (the ERP) and a source location plus projection pattern for this signal, we can now combine these into a single component. Components again are represented as structure arrays in this toolbox, with separate fields for the component's source location, orientation, and its activation signal.
c = struct();
c.source = source; % obtained from the lead field, as above
c.signal = {erp}; % ERP class, defined above
c = utl_check_component(c, lf);Note that, just as for classes, there is a function, utl_check_component, to validate the component structure and fill in any missing parameters. For example, if no orientation is indicated, this function reverts the source's orientation to a default value obtained from the lead field.
(Also have a look at utl_create_component in the examples further below for a shorthand function to replace manually assigning structure arrays with signal and source fields. It also automatically verifies the resulting components.)
If desired, changing a source's orientation also happens here, at the level of components. It can be done simply by changing the value in the orientation field of the corresponding component. You can input manual values, or see the above section on orientation for functions to obtain different orientations.
c.orientation = [0 1 0];
c.orientation = utl_get_orientation_pseudoperpendicular(source, lf);We now have the minimum we need to simulate scalp data. Scalp data is simulated by projecting all components' signal activations through their respective, oriented source projections, and summing all projections together. Right now, our scalp data would contain the activation of only one component, with one single and fixed activation pattern coming from one and the same location.
scalpdata = generate_scalpdata(c, lf, epochs);After the above step, scalpdata contains the channels-by-samples-by-epochs simulated data, but no additional information, such as time stamps, channel names, et cetera.
We can turn this into a dataset according to the EEGLAB format, which has a standard structure to save such information. Doing so will also allow us to use EEGLAB's analysis tools, for example, to see the scalp projection time course of the ERP we just simulated. At this point, two optional parameters in the configuration array epochs are taken into account.
epochs.marker = 'event 1'; % the epochs' time-locking event marker
epochs.prestim = 200; % pre-stimulus period in ms. note that this
% only affects the time indicated in the final
% dataset; it is ignored during simulation.
% i.e., a simulated latency of 500 ms becomes a
% latency of 300 ms when a prestimulus period
% of 200 ms is later applied. you can use
% utl_shift_latency to shift all latencies in a
% class to fit the pre-stimulus period.
EEG = utl_create_eeglabdataset(scalpdata, epochs, lf);
pop_topoplot(EEG, 1, [100, 200, 250, 300, 350, 400, 500], '', [1 8]);The above code named the simulated event event 1, and set its zero point at 200 ms into the epoch. utl_create_eeglabdataset takes the simulated data, the configuration array, and the lead field and compiles an EEGLAB-compatible dataset. The call to pop_topoplot demonstrates this compatibility.
To save the dataset in EEGLAB format, use EEGLAB's function pop_saveset.
Keep in mind that EEGLAB uses the variable EEG internally to refer to the currently active dataset. Therefore, if you have assigned the dataset to the variable EEG, calling eeglab redraw redraws the main menu and recognies the newly created or updated variable as an active dataset. You can then use EEGLAB's GUI to further work with the data.
If we scroll through the data, we see that all 100 epochs we generated are exactly the same, having a peak at exactly the indicated latency with a width of exactly 200.
pop_eegplot(EEG, 1, 1, 1);This is of course unrealistic and defeats the entire purpose of even simulating multiple epochs in the first place.
We can add variability to the signal by indicating allowed random deviations or shifts as well as specific slopes. A deviation of 50 ms for our peak latency allows this latency to vary +/- 50 ms between trials, following a normal distribution with the indicated deviation being the six-sigma range, capped to never exceed the indicated maximum.
erp.peakLatencyDv = 50;
erp.peakAmplitudeDv = .2;
c.signal = {erp};
EEG = utl_create_eeglabdataset(generate_scalpdata(c, lf, epochs), ...
epochs, lf);
pop_eegplot(EEG, 1, 1, 1);When multiple peaks are defined, note that peakLatencyDv applies to each peak latency value separately. Another parameter, peakLatencyShift, works the same way but applies to all values equally.
Indicating a slope results in a consistent change over time. An amplitude of 1 and an amplitude slope of -.75, for example, results in the signal having a peak amplitude of 1 in the first epoch, and .25 in the last.
erp.peakAmplitudeSlope = -.75;
c.signal = {erp};
EEG = utl_create_eeglabdataset(generate_scalpdata(c, lf, epochs), ...
epochs, lf);
figure; pop_erpimage(EEG, 1, [25], [[]], 'Pz', 10, 1, {}, [], '', ...
'yerplabel', '\muV', 'erp', 'on', 'cbar', 'on', ...
'topo', {[25] EEG.chanlocs EEG.chaninfo});After these changes, the possible shape that the ERP can take varies significantly. We can plot the extreme values in one figure. In blue: extreme values for the first epoch; in red: extreme values for the last.
plot_signal_fromclass(erp, epochs);It is also possible to not have the signal occur every epoch. Instead, a probability can be indicated varying between 1 (always occurs) and 0 (never occurs). This probability can have a probabilitySlope as well, making the signal more or less likely to occur over time.
See utl_set_dvslope for a convenience function to set all deviation and/or slope parameters of a class to a specific value relative to the parameters' base values.
Instead of an ERP, we can also project a different signal onto the scalp. For example, we can project noise i.e., a (pseudo)random signal with certain spectral features. We can define a new noise activation class that generates brown noise of a given maximum amplitude.
noise = struct( ...
'type', 'noise', ...
'color', 'brown', ...
'amplitude', 1);
noise = utl_check_class(noise);
c.signal = {noise};
EEG = utl_create_eeglabdataset(generate_scalpdata(c, lf, epochs), ...
epochs, lf);
pop_eegplot(EEG, 1, 1, 1);It does not have to be either/or. We can also add this noise and the already-existing ERP activation together. We can do this by simply adding both the ERP class variable, and the noise class variable to the component's signal field.
c.signal = {erp, noise};
EEG = utl_create_eeglabdataset(generate_scalpdata(c, lf, epochs), ...
epochs, lf);
pop_eegplot(EEG, 1, 1, 1);When a component's signal activity is simulated, all of its signals are simulated separately and then summed together before being projected through the lead field. It is thus possible to generate any number of signal activation patterns from the same source location using a single component, for example, as we did here, in order to add noise to an otherwise clean signal. Also see utl_add_signal_tocomponent to add an additional signal activation class to an existing component.
The noise class can simulate different types of coloured noise: white, pink, brown, blue and purple. By default, these are generated from a Gaussian process using the DSP toolbox. A third-party script, not using the DSP toolbox, allows coloured noise to be generated using a uniform process as well, e.g. white-unif, brown-unif, et cetera.
In EEG analyses, we may be used to speak of 'sources' in the brain to indicate specific areas that we localised on the basis of specific activity coming from that area. In such contexts, the concept of 'source' thus combines a number of different pieces of information. For SEREEGA, the word 'source' refers to the sources as they are represented in a lead field. They can be combined with additional information, primarily the signal, to form components. It is thus at the level of the component where the building blocks of the simulation are defined, by the combination of signal activations, source locations, and their orientations.
A component must have at least one signal to project, but can have multiple signals, as we said above. When a component's activation is simulated, all of its signals are simulated separately and summed together before being projected onto the scalp.
A component must have at least one source location, but can also have multiple. When one and the same component contains multiple sources, that component's signal activation is simulated once per epoch, and then projected as such from all indicated sources. This can be used to simulate bilateral sources, for example. When multiple sources are indicated, they all also have their own orientation. (The section on component variability further below discusses an alternative way to use multiple sources in the same component. This is no longer the default behaviour of SEREEGA, but can still be used.)
You can define as many components as you want using any combination of sources and signals. The first argument of generate_scalpdata is a struct containing any nonzero number of components.
Keep in mind that adding multiple sources to one and the same component is completely different from adding the same signal(s) to multiple different components that each have one source. In the former case, the exact same signal activation is projected simultaneously from all sources. In the latter case, separately simulated instances of the signal activation are projected from each source.
This, then, results in the following simplified workflow using SEREEGA:
We have seen the event-related potential (ERP) and noise types of signal activations above. SEREEGA supports additional signal types, and can easily be extended to support more. The next few sections will address the other available signals.
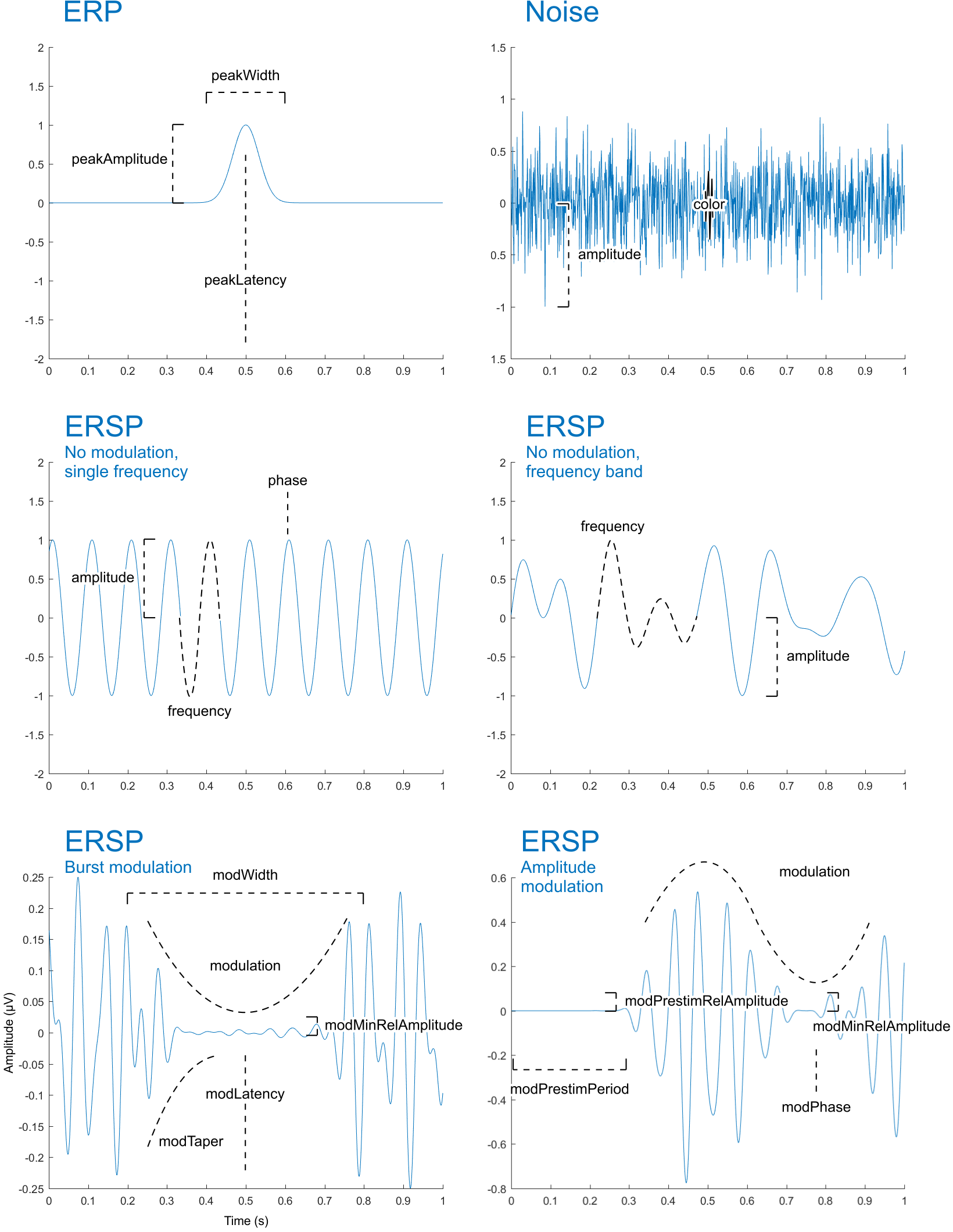
Although not all parameters can easily be visualised, the below figure provides an illustration of three base signal types (ERP, noise, and ERSP, described next) along with its main parameters.
ERP and noise classes are examples of two types of signal activations. A third type concerns oscillatory activations. In its basic form, it is merely a sine wave of a given frequency, amplitude and, optionally, phase.
ersp = struct( ...
'type', 'ersp', ...
'frequency', 20, ...
'amplitude', .25, ...
'modulation', 'none');
ersp = utl_check_class(ersp);
plot_signal_fromclass(ersp, epochs);Alternatively, it can be a broadband signal. In this case, instead of indicating a single frequency, a sequence of band edge frequencies is given. For example, the frequency specification below will result in a signal with maximum spectral power between 15 and 25 Hz, with transitions between 12-15 and 25-28 Hz. This is generated by filtering uniform white noise in the indicated frequency band. In case a frequency band is indicated, phase will be ignored.
ersp = struct( ...
'type', 'ersp', ...
'frequency', [12 15 25 28], ...
'amplitude', .25, ...
'modulation', 'none');
ersp = utl_check_class(ersp);
plot_signal_fromclass(ersp, epochs);Note that the above two examples contained a modulation field that was set to none. When this field is set to different values, these signals serve as base oscillatory signals that are then modulated in the indicated way.
The base oscillatory signal can be modulated such that it appears only as a single frequency burst, with a given peak (centre) latency, width, and taper.
ersp.modulation = 'burst';
ersp.modLatency = 500; % centre of the burst, in ms
ersp.modWidth = 100; % width (half duration) of the burst, in ms
ersp.modTaper = 0.5; % taper of the burst
ersp = utl_check_class(ersp);
plot_signal_fromclass(ersp, epochs);The inverse of the above is also possible. It results in an attenuation in the given window. In both cases, it is also possible to set a minimum amplitude, in order to restrict the attenuation, which is otherwise 100%.
ersp.modulation = 'invburst';
ersp.modWidth = 300;
ersp.modMinRelAmplitude = 0.05;
plot_signal_fromclass(ersp, epochs);This allows the base signal's amplitude to be modulated according to the phase of another. In the example below, a 20 Hz base frequency is modulated using a 2 Hz wave.
ersp.frequency = 20;
ersp.modulation = 'ampmod';
ersp.modFrequency = 2;
ersp.modPhase = .25;
ersp = utl_check_class(ersp);
plot_signal_fromclass(ersp, epochs);Indicating the phase of the modulating wave is optional, as is indicating the modMinRelAmplitude, as above. Additionally, the amplitude-modulated signal can be attenuated completely during a given baseline period, using a tapering window similar to the one used for frequency bursts.
ersp.frequency = [12 15 25 28];
ersp.modPrestimPeriod = epochs.prestim;
ersp.modPrestimTaper = .5;
plot_signal_fromclass(ersp, epochs);At this point, we may again put all these three activation classes together in our previously-defined component, and see the resulting EEG:
c.signal = {erp, ersp, noise};
EEG = utl_create_eeglabdataset(generate_scalpdata(c, lf, epochs), ...
epochs, lf);
pop_eegplot(EEG, 1, 1, 1);Note that variability parameters (deviation, slope) can be added to almost all parameters defining ERSP signals, as above with ERP parameters.
Most SEREEGA activation classes are intended for the procedural generation of an activation signal based on given parameters. To include time series that were externally obtained, generated, or modulated, a data class is provided. This allows a signal activation to be extracted from a matrix containing time series, based on given indices.
% creating matrix of specific random data
s = rng(0, 'v5uniform');
randomdata = randn(epochs.n, epochs.srate*epochs.length/1000);
rng(s);
% adding generated random data to data class
data = struct();
data.data = randomdata;
data.index = {'e', ':'};
data.amplitude = .5;
data.amplitudeType = 'relative';
data = utl_check_class(data, 'type', 'data');
plot_signal_fromclass(data, epochs);This class projects, for each simulated epoch e, the eth row of data from the matrix randomdata. plot_signal simply plots the first epoch.
When used as a single signal, an autoregressive model (ARM) class generates a time series where each sample depends linearly on its preceding samples plus a stochastic term. The order of the model, i.e. the number preceding samples the current sample depends on, can be configured. The exact model is generated using random parameters and fixed for that class upon verification.
For example:
% obtain and plot an ARM signal of order 10
arm = struct('order', 10, 'amplitude', 1);
arm = arm_check_class(arm);
plot_signal_fromclass(arm, epochs);(Note that, alternative to calling utl_check_class and indicating the class type, or indicating it in the class structure itself, we can also call the class's own check function directly. This is in fact what utl_check_class does after having determined the class type.)
The strength of autoregressive modelling however is in its ability to include interactions between time series. Thus, it is also possible to simulate connectivity between multiple source activations. Uni- and birectional interactions can be indicated such that a time-delayed influence of one source on another is included in the signal. Either the number of interactions is configured and the toolbox will randomly select the interactions, or a manual configuration of the exact directionalities can be indicated. The model order is the same for all interactions.
Interacting signals are not simulated at runtime, but generated beforehand and included in the simulation as data classes. For example:
% obtaining two data classes containing ARM-generated signals: two time
% series series with one unidirectional interaction between them, and a
% model order of 10
arm = arm_get_class_interacting(2, 10, 1, epochs, 1);
% getting two components with sources that are 100 mm apart
armsourcelocs = lf_get_source_spaced(lf, 2, 100);
armcomps = utl_create_component(armsourcelocs, arm, lf);
plot_component(armcomps, epochs, lf);When a large number of classes are needed, it is cumbersome to define them all by hand. When the exact parameter details do not matter, it is possible to use random class generation methods. Both ERP and ERSP classes have methods to obtain a number of classes, randomly configured based on allowed parameter ranges.
For example:
% obtain 10 random source locations, at least 5 cm apart
sourcelocs = lf_get_source_spaced(leadfield, 10, 50);
% obtain 64 random ERP activation classes. each class will have
% either 1, 2, or 3 peaks, each centred between 200 and 1000 ms,
% widths between 25 and 200 ms, and amplitudes between -1 and 1.
erp = erp_get_class_random([1:3], [200:1000], [25:200], ...
[-1:.1:-.5, .5:.1:1], 'numClasses', 10);
% combining into components
c = utl_create_component(sourcelocs, erp, lf);
% plotting each component's projection and activation
plot_component(c, epochs, lf);The generate_scalpdata function returns both a summed channels x samples x epochs array of simulated scalp data, and a components x samples x epochs array of the individual component source activations.
Since we know the exact projection of each component, it is possible to generate ground-truth ICA mixing and unmixing matrices. This is done by concatenating the components' (mean) projection patterns, and taking its inverse.
% obtaining 64 random source locations, at least 2.5 cm apart
sourcelocs = lf_get_source_spaced(leadfield, 64, 25);
% each source will get the same type of activation: brown coloured noise
signal = struct('type', 'noise', 'color', 'brown', 'amplitude', 1);
% packing source locations and activations together into components
components = utl_create_component(sourcelocs, signal, lf);
% obtaining mixing and unmixing matrices
[w, winv] = utl_get_icaweights(components, leadfield);In case an EEGLAB dataset has been created from generated scalp data, e.g. using utl_create_eeglabdataset, the convenience function utl_add_icaweights_toeeglabdataset can be used to add the weights directly to the dataset.
Just as activation signals can have an epoch-to-epoch variability, so too can the location and orientation of the signal's source vary. The orientationDv field of the component struct contains an n-by-3 matrix containing the maximum permissible deviation for the orientation of each of the n sources indicated for that component.
Indicating more than one source for a component, as mentioned above, results in a signal projected from all those sources. However, in versions of SEREEGA older than v1.1.0, this was interpreted as location variability of the source. For each epoch, it randomly selected one of the indicated sources to project the signal activation from. This behaviour can be reinstated using the legacy_rndmultsrc argument when calling generate_scalpdata. As such, you could use lf_get_source_inradius to get all sources around a particular point, and add all of these to a single component to simulate activity coming from variable locations in the same brain region.
It is relevant to highlight that the various lead fields supported by SEREEGA can have different units of measure. This is the case, for example, for all three of the originally supported lead fields: NYHead, FieldTrip, and Pediatric Head Atlas. Unfortunately, although knowing the units is important to interpret the results correctly, these have not been reported in the corresponding literature. Consequently, the units of the result are relative to the leadfield employed. As a workaround, the generate_scalpdata function contains a normaliseLeadfield argument, which normalises the leadfield values to at least maintain comparability.
An exception to this is the lead field converted from Brainstorm. Brainstorm explicitly utilises the International System; thus, every leadfield generated with it is expressed in lf_generate_frombrainstorm automatically converts this to
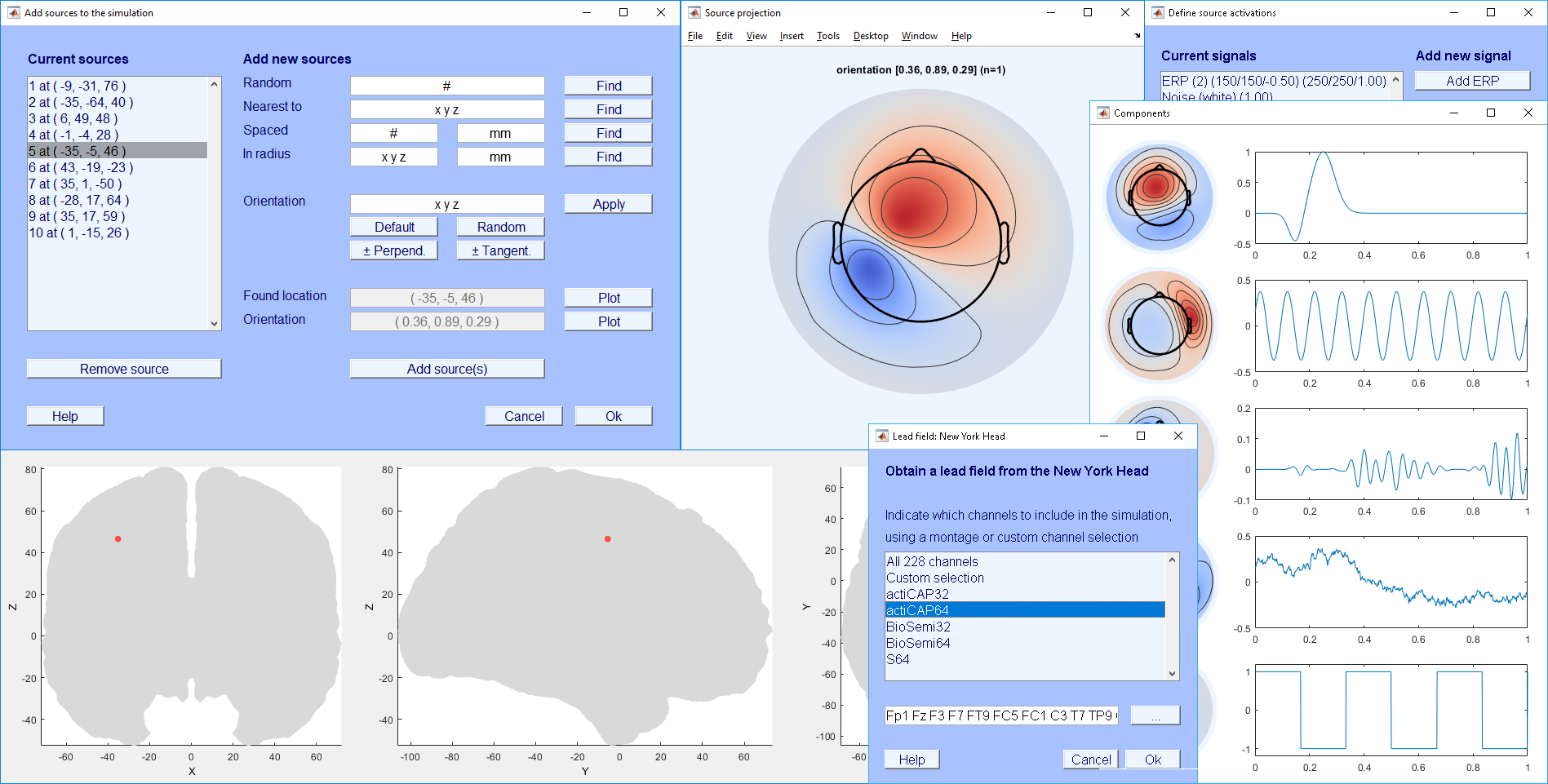
When using SEREEGA as an EEGLAB plug-in, it will appear as a separate sub-menu in EEGLAB's "Tools" menu. The SEREEGA menu provides options to follow the basic steps outlined before in this tutorial.
New simulation loads a new, empty EEGLAB dataset. Since all SEREEGA-related information is saved as part of a dataset, SEREEGA needs a dataset to be loaded. Saved information includes the epoch configuration, lead field, source positions, signals, and components. It stores this in EEG.etc.sereega. When finally data is simulated, it reads this information and fills the placeholder EEGLAB dataset with the simulated data.
The option Configure epochs allows you to indicate how much data is to be simulated. The various options in the Configure lead field sub-menu allow you to add one of these lead fields to the dataset, thus making it the basis for the simulation. Note that these options can all be changed at any time, but they must be set before the following functions can be used.
The Configure components sub-menu contains three options which can be used to define the components that will underly the simulated data.
First, Select source locations provides a dialog window that allows you to find random or specific sources in the brain (i.e. in the configured lead field), determine their desired orientation, and add them to the simulation. Two plots can support you in this task: one for the source's location, and one for its projection. If you have found a source you wish to keep, click Add source(s) to add it to the list at the left, and OK to save the list to the dataset.
Define signal activations, like the previous window to select source locations, provides a list of signals currently stored in the dataset, and options to add additional signal classes. Each signal type has its own button, which pops up a second window to input the parameters that define each signal. Values that are indicated with an asterisk (*) in both their row and column are required. Click OK to save the list.
Now it must be decided which of the defined signals will project from which of the selected source locations. The Assign signals to sources dialog shows a list of the sources selected earlier, and allows you to assign the defined signal classes to these, thus completing the definition of components.
Finally, Simulate data simulates the data and populates the EEGLAB dataset with the corresponding values.
Before saving the generated dataset, you may want to use the Remove/add lead field data option in the Misc menu. Here, you can remove the lead field from the dataset to save space. The lead field can always be re-generated later.
SEREEGA was designed to make it relatively easy to add new lead fields from different sources and new signal activation patterns.
For general changes and extensions, please use the existing scripts as examples. The general function name pattern is [prefix_]action_object[_clarification], with the optional prefix referring to a class of connected or otherwise related functions (such as erp_ for scripts related to the ERP signal type, plot_ for plotting functions, etc.), or utl_ for the class of miscellaneous utilities.
This project adheres to Semantic Versioning. SEREEGA's modularity means that extensions as described below should result in patch or minor version increments; no API changes should be necessary.
Files and scripts related to lead fields in general are in the ./leadfield directory and prefixed with lf_. Files and scripts related to specific lead fields are in accordingly named subdirectories of ./leadfield. The ./leadfield/nyhead directory is a good example. There is one file that contains the lead field data, one file containing the channel locations in an EEGLAB-readable format, and a script to generate a lead field from these files.
To add a lead field to SEREEGA, create a lf_generate_<yourleadfield>.m script that outputs the lead field in the given format:
% a lead field structure array containing the follow fields
% .leadfield - the lead field, containing projections in three
% directions (xyz) for each source, in a
% nchannels x nsources x 3 matrix
% .orientation - a default orientation for each soure, or
% zeros(nsources, 3) if no meaning default is available
% .pos - nsources x 3 xyz MNI coordinates of each source
% .chanlocs - channel information in EEGLAB format
% .atlas - nsources x 1 cell of strings containing the atlas,
% i.e. indciating the name of the corresponding region
% for each sourceIf the lead field does not come with a meaningful default orientation, set the defaults to all zeros to force a manual orientation.
Each entry in the atlas must start with one of the three generic categories that SEREEGA recognises: Brain, Eye, or Muscle (not case sensitive). If the lead field does not come with a meaningful atlas, simply only indicate one of these three. Use utl_sanitize_atlas to make sure the final atlas format is compatible with other SEREEGA functions.
Note that SEREEGA's standard plotting functions calculate a brain boundary based on dipole locations. The accuracy of these plots thus depend on the resolution of the lead field. Alternatively, there are two _dipplot plotting functions functions which use a standard head model as backdrop.
Try to make your newly added lead field adhere to the standard coordinates if possible. See lf_generate_frompha for an example where the head model's coordinates are transformed to approximately fit the standard MNI head model. This is the head model that is used as backdrop by the plot_chanlocs_dipplot and plot_source_location_dipplot functions, which can be used as reference.
Some head models, such as the Pediatric Head Atlas of ages up to 2 years old, will obviously not fit the standard, adult MNI model. If none of SEREEGA's plotting functions seem to fit your head model, you could provide separate plotting functions for your lead field. These should then be in the lead field's own directory and named accordingly.
The electrode coordinates require special attention. These should be centred around a similar zero-point as the standard MNI head midpoint, in order for EEGLAB's topoplot to be able to properly plot the lead field's projection patterns. Note that EEGLAB's readlocs, which is used by SEREEGA, reads the electrode positions with X=Y and Y=-X.
(If you came to this section of the documentation because you wanted to add a new source to an existing lead field in your work space: This can be done manually using lf_add_source.)
Files and scripts related to signal activation classes are in class-specific subdirectories of the ./signal directory. To add new classes of signal activations to SEREEGA, the following files, containing functions of the same name, must be supplied in a new subdirectory. denotes the name of the new class.
<class>_check_class - Takes a class definition structure, verifies/completes it to pass the requirements of the new class, and returns a class variable that is compatible with all other functions. The class must have a 'type' field that is equal to , allowing utl_check_class to find this file. This file is also where the class documentation should be provided. If any deviation and slope fields are provided, add 'Dv' or 'Slope' to the base field name. A slope for a 'frequency' field should thus be called 'frequencySlope', not e.g. 'freqSlope'. This allows e.g. utl_apply_dvslope to function properly. Similarly, if any latencies are to be indicated, use a field name that ends in Latency and describes what latency it is, as e.g. with peakLatency for ERPs and modLatency for ERSP modulations. This allows e.g. utl_shift_latency to function properly.
<class>_generate_signal_fromclass - Takes a (verified) class structure, an epochs configuration structure, and (at least accepts) an epochNumber argument (indicating the number of the currently generated epoch) and 'baseonly' (whether or not to ignore the deviations, slopes, et cetera). Returns a 1-by-nsamples signal activation time course.
<class>_plot_signal_fromclass - Takes a (verified) class structure, an epochs configuration structure, and (at least) accepts the optional
'newfig' (to open a new figure) and 'baseonly' (whether or not to also plot possible signal variations) arguments. This plots a/the signal activation and returns the figure handle if a new figure was opened.
For inclusion into the GUI, the following files are additionally needed:
<class>_class2string - Takes a verified class structure and returns a string describing it. This is used to represent signal classes in the GUI. For example, an ERP with a single 200-ms wide peak at latency 100 and an amplitude of 3, is represented as ERP (1) (100/200/3.00).
pop_sereega_add<class> - Takes an EEGLAB dataset, provides a dialog to define the parameters of the class in question, and adds it to EEG.etc.sereega.signals. Furthermore, pop_sereega_signals needs to be adapted, by adding an extra button to add this type of signal.
The function utl_get_component_fromtemplate contains a number of predefined components, complete with source locations, activation signals, and orientations. For example, utl_get_component_fromtemplate('p300_erp', lf) returns two components together simulating a generic P300 activation pattern.
If you construct a new component for your project that may be of use to others as well, please feel free to extend the list of templates and share it with the project. Make sure it is independent of the lead field used (e.g. do not rely on default orientations but make them explicit).
Of course, do not uncritically rely on these templates. Even when proper care is taken, their parameters may still have been set to suit a very different purpose than the one you may have in mind when using them.
The function utl_get_montage contains a number of predefined montages, i.e., collections of channel labels representing specific cap layouts (for example a standard BioSemi cap with 32 electrodes, a Brain Products EASYCAP with 64 channels, et cetera). This file can simply be extended by adding additional cases, representing the montage name, defining the respective labels in a cell array.
A quick sample script is given in the introduction. This section contains a few other, not necessarily complete, examples.
Rather than manually defining each class, an anonymous function such as the one below can provide more control than e.g. erp_get_class_random. This example also shows a quick way to simulate two different conditions in a single data set.
% anonymous function to quickly generate valid ERP classes
% of specified latency and amplitude, with a fixed peakWidth,
% and fixed-but-relative deviation for all values
erp = @(lat,amp) ...
(utl_set_dvslope( ...
utl_check_class(struct( ...
'type', 'erp', ...
'peakLatency', lat, ...
'peakWidth', 200, ...
'peakAmplitude', amp)), ...
'dv', .2));
noise = utl_check_class(struct( ...
'type', 'noise', ...
'color', 'brown', ...
'amplitude', 1));
% getting 64 noise components, same for each condition
sources = lf_get_source_spaced(lf, 64, 25);
[comps1, comps2] = deal(utl_create_component(sources, noise, lf));
% now differentiating between conditions:
% adding ERP to first component of first condition, and
% adding different ERP to same component of second condition
comps1(1) = utl_add_signal_tocomponent(erp(300, 1), comps1(1));
comps2(1) = utl_add_signal_tocomponent(erp(500, -.5), comps2(1));
% simulating data, converting to EEGLAB, and reorganising
data1 = generate_scalpdata(comps1, lf, epochs);
data2 = generate_scalpdata(comps2, lf, epochs);
EEG1 = utl_create_eeglabdataset(data1, epochs, lf, 'marker', 'event1');
EEG2 = utl_create_eeglabdataset(data2, epochs, lf, 'marker', 'event2');
EEG = utl_reorder_eeglabdataset(pop_mergeset(EEG1, EEG2));The following code roughly mimics the connectivity benchmarking simulation framework proposed by Haufe and Ewald (2016). Note that the signal-to-noise ratio in this case is determined directly by the amplitudes of the ARM and noise classes. Further down in this section there is sample code covering how to specify the signal-to-noise ratio manually.
% getting two ARM components with one unidirectional interaction,
% at least 10 cm apart
arm = arm_get_class_interacting(2, 10, 1, epochs, 10);
armsourcelocs = lf_get_source_spaced(lf, 2, 100);
armcomps = utl_create_component(armsourcelocs, arm, lf);
% getting 500 pink noise components
noise = struct('type', 'noise', 'color', 'pink', 'amplitude', 1);
noisesourcelocs = lf_get_source_random(lf, 500);
noisecomps = utl_create_component(noisesourcelocs, noise, lf);
% simulating data
data = generate_scalpdata([armcomps, noisecomps], lf, epochs);
EEG = utl_create_eeglabdataset(data, epochs, lf);In SEREEGA we define all signals at the source level, and this includes noise activations which are added to components just as other signals are. In this fashion, it can be difficult to control the exact signal-to-noise ratio (SNR) at the scalp, as this depends on the projections and varies with the number of sources. Instead, it is possible to simulate scalp data for signal and noise aspects separately, and mix them together using a given SNR using utl_mix_data.
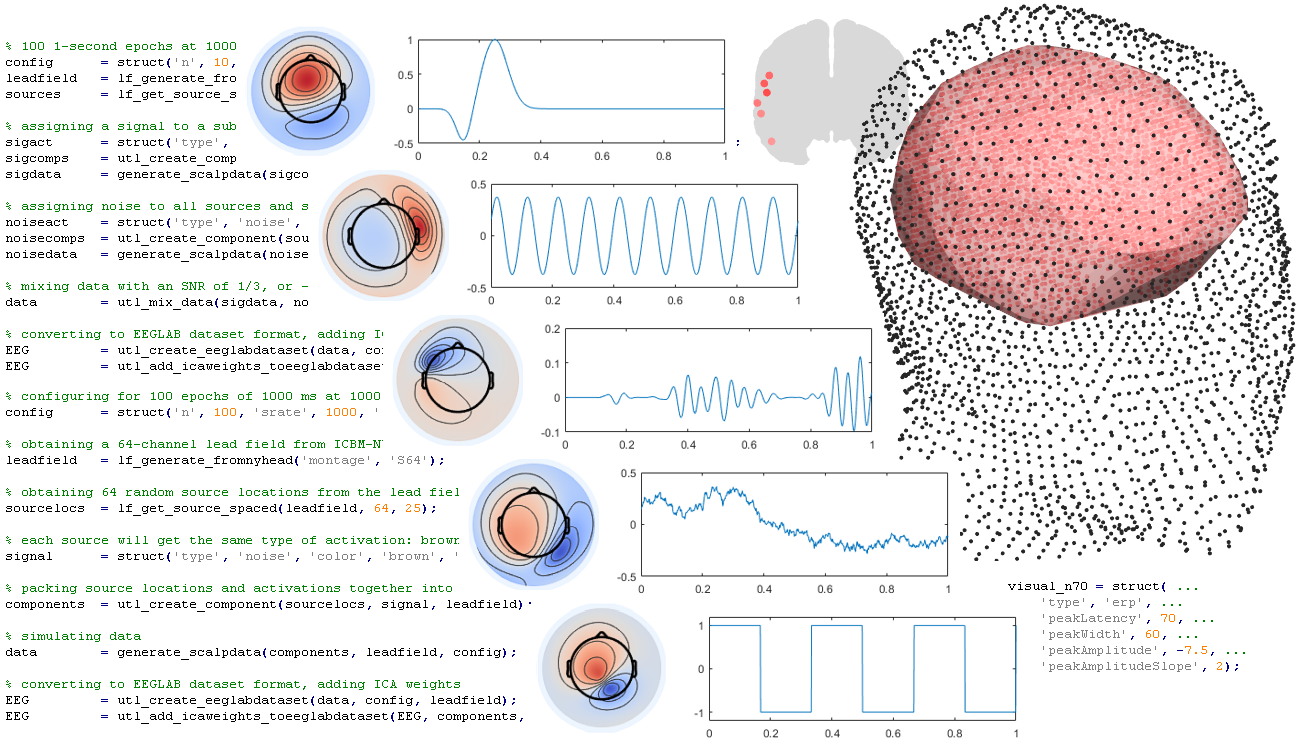
% obtaining 64 source locations
sources = lf_get_source_spaced(leadfield, 64, 25);
% assigning a signal to a subset of the sources and simulating data
sigact = struct('type', 'ersp', ...
'frequency', 10, 'amplitude', 1, 'modulation', 'none');
sigcomps = utl_create_component(sources(1), sigact, leadfield);
sigdata = generate_scalpdata(sigcomps, leadfield, config);
% assigning noise to all sources and simulating data
noiseact = struct('type', 'noise', 'color', 'brown', 'amplitude', 1);
noisecomps = utl_create_component(sources, noiseact, leadfield);
noisedata = generate_scalpdata(noisecomps, leadfield, config);
% mixing data with an SNR of 1/3, or -6 dB
data = utl_mix_data(sigdata, noisedata, 1/3);Also note that an additional source of noise, sensor noise, can be added to generated scalp data using the sensorNoise argument of generate_scalpdata. This noise has no dependencies across channels or samples.
Feel free to contact me at [email protected].
Fabien Lotte provided two early drafts of what are now lf_generate_fromfieldtrip and erp_generate_signal. I'd like to thank Mahta Mousavi for the band-pass filter design, Stefan Haufe for the autoregressive signal generation code and accompanying support, and Ramón Martínez-Cancino for his assistance with the GUI development. Part of this work was supported by the Deutsche Forschungsgemeinschaft (grant number ZA 821/3-1), Idex Bordeaux and LabEX CPU/SysNum, and the European Research Council with the BrainConquest project (grant number ERC-2016-STG-714567), and Volkswagen Foundation.