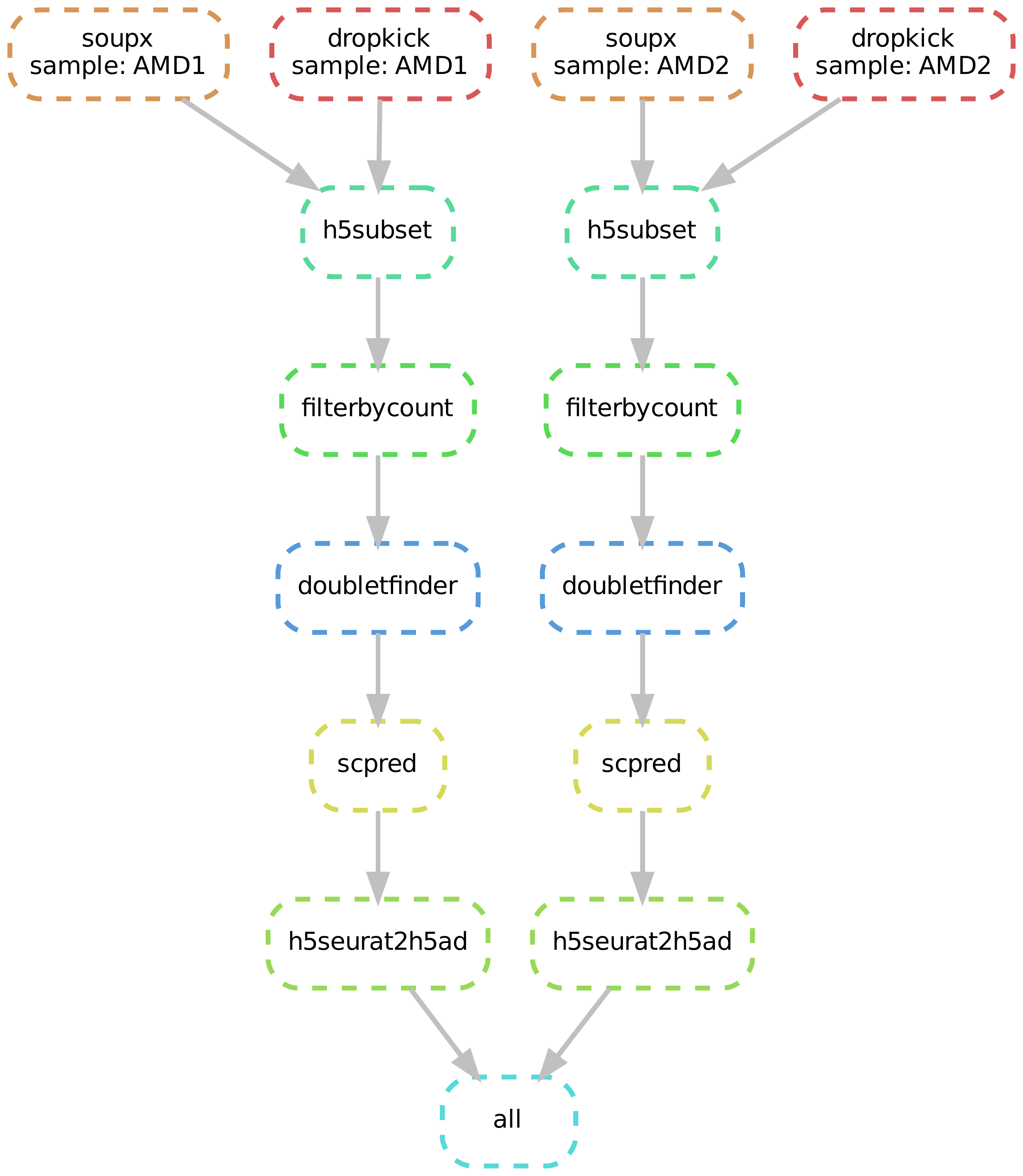
Cellqc standardizes the qualiy control of single-cell RNA-Seq (scRNA) data to render clean feature count matrices from Cell Ranger outputs. Cellqc is implemented using the Snakemake workflow management system to enhance reproduciblity and scalablity of data analysis. Briefly, the QC pipeline starts from raw count feature matrices from Cell Ranger. Dropkick filters out predicted empty droplets, and SoupX purify the transcriptome measurement by substracting the background trancripts. DoubletFinder further detects the potential doublets and retain clean count feature matrices for singlets. Cell types are annotated for clean cells by a reference database using scPred.
It is easy to install cellqc via conda at https://anaconda.org/bioconda/cellqc. To use the full function of cellqc, please also install several dependencies outside conda. It is encouraged to use the C++ implementation mamba to speed up the installation. E.g.,
conda config --add channels defaults --add channels bioconda --add channels conda-forge
mamba create -n cellqc cellqc python=3.10
conda activate cellqc
# Downgrade Seurat for seurat-disk, as Seurat v5 is not supported in SeuratDisk. A new Seurat is not necessary.
mamba install r-seurat=4 r-seuratobject=4 r-matrix=1.6.1
# Build from source
Rscript -e "remotes::install_github(c('mojaveazure/seurat-disk', 'immunogenomics/harmony', 'powellgenomicslab/scPred'), upgrade=F)"
# Bug fix @counts for Seurat object, instead of chris-mcginnis-ucsf/DoubletFinder
Rscript -e "remotes::install_github('lijinbio/DoubletFinder', upgrade=F, force=T)"
# Downgrade dependencies for dropkick
mamba install numpy=1.21 scanpy=1.9.1 matplotlib=3.6 anndata=0.7.8 pandas=1.5
pip install dropkick
pip install -U cellqc # to install the latest version
Dependent software are summarized below.
To test the installation, simply run
cellqc -h
Cellqc requires a sample file for sample information and an optional configuration file for pipeline parameters.
-
The sample file (e.g.,
samples.txt) is a tab-delimited file with headers:sample,cellranger, and/ornreaction.- The
samplecolumn is the sample ID per sample. - The
cellrangeris the Cell Ranger output directory. See Cell Ranger Outputs for an example directory. - The optional third column
nreactionis the number of reactions in the library preparation, which is useful to infer expected doublets for a sample with a Cell Ranger analysis using combined raw reads from multiple reactions. If thenreactioncolumn is not specified in the sample file, the default 1 reaction is used for all samples.
- The
-
A configuration file is in the YAML format. It is optional. The default parameters can be used as below. See the next section for the inspection of configuration.
dropkick:
skip: false
method: multiotsu
numthreads: 1
filterbycount:
mincount: 500
minfeature: 300
mito: 10
doubletfinder:
skip: false
findpK: false
numthreads: 5
pK: 0.01
scpred:
skip: true
reference: /path_to_reference/scPred_trainmodel_RNA_svmRadialWeights_scpred.rds
threshold: 0.9
The configuration file is in a YAML format. An example configuration can be found at the example directory.
- dropkick
This section defines parameters for empty droplet removal by dropkick.
| Parameter | Description |
|---|---|
| dropkick.skip | Skip Dropkick and use the estimated cells from Cell Ranger alone (using EmptyDrops) if set true. If set false, to estimate further empty droplets by Dropkick. Be cautious that Dropdick might predict a significant number of false negatives for a poor library. |
| dropkick.method | The thresholding method for labeling the training data for true cells, such as multiotsu, otsu, li, or mean. |
| dropkick.numthreads | Number of threads. Dropkick will use significant memory. One thread is suggested for this step. |
- filterbycount
To filter cells by nCount, nFeature, and percentage of mitochondria reads.
| Parameter | Description |
|---|---|
| filterbycount.mincount | Minimum counts for a cell. |
| filterbycount.minfeature | Minimum features for a cell. |
| filterbycount.mito | Maximum percentage of mitocondria transcripts. |
- doubletfinder
This section includes three parameters for doublet removal by DoubletFinder.
| Parameter | Description |
|---|---|
| doubletfinder.skip | Skip doublet detection and removal. |
| doubletfinder.findpK | To estimate the neighbor size (pK) by mean-variance bimodality coefficients if true. If set false, skip the estimation and use the preset pK value. |
| doubletfinder.numthreads | Number of threads. |
| doubletfinder.pK | A preset neighbor size (pK). Will be used if doubletfinder.findpK=false. |
- scpred
A pre-trained classifier for cell-type annotation by scPred.
| Parameter | Description |
|---|---|
| scpred.skip | Skip the automated cell type prediction by scPred if true. This is useful for a sample without a pre-trained reference. |
| scpred.reference | The pre-trained reference classifier saved in a RDS file. See https://github.com/powellgenomicslab/scPred |
| scpred.threshold | Threshold for a positive prediction. |
Three result files are generated under a result subdirectory. result/*.h5seurat and result/*.h5ad files are count matrices after processing with QC metrics such as "pANN" for proportion of artificial nearest neighbors, and/or "scpred_prediction" for predicted cell type. A report file result/report.html is a summary of QC metrics. A postproc subdirectory with postproc/*.h5ad files is also generated for basic post-processing. This includes adding a prefix to the cell barcode, ensuring unique variable names, and cleaning the raw layer from the .h5ad file.
This example demonstrates the pipeline on two AMD samples. The test data consists of Cell Ranger output directories of two AMD samples, as well as a pretrained calssifier for cell-type annotation.
https://bcm.box.com/s/nnlmgxh8avagje93cih20g1dsxx14if4
After feeding the file locations, a sample file (e.g., samples.txt) looks like below.
sample cellranger
AMD1 /path/to/cellranger/AMD1/outs
AMD2 /path/to/cellranger/AMD2/outsBelow command is to run the pipeline by the installed entrypoint cellqc.
cellqc -d "$outdir" -t 8 -- samples.txt # with default parameters
cellqc -d "$outdir" -t 8 -c config.yaml -- samples.txt # by customized parameters in config.yaml
A directed acyclic graph (DAG) of jobs will be generated. For example,
A report of result files will be also produced, such as report.html.